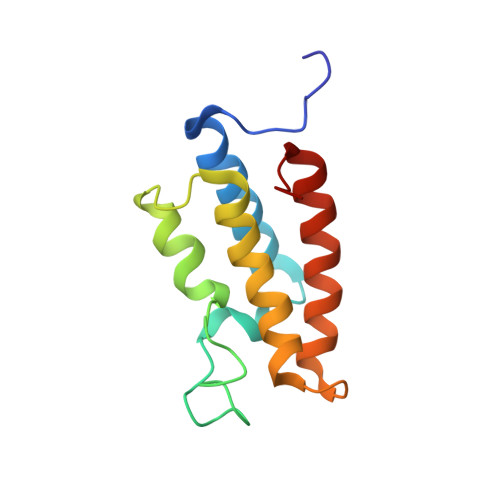
Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
Zeng, L., Li, J., Muller, M., Yan, S., Mujtaba, S., Pan, C., Wang, Z., Zhou, M.M.(2005) J Am Chem Soc 127: 2376-2377
- PubMed: 15724976
- DOI: https://doi.org/10.1021/ja044885g
- Primary Citation of Related Structures:
1WUG, 1WUM - PubMed Abstract:
Development of drug resistance from mutations in the targeted viral proteins leads to continuation of viral production by chronically infected cells, contributing to HIV-mediated immune dysfunction. Targeting a host cell protein essential for viral reproduction, rather than a viral protein, may minimize the viral drug resistance problem as observed with HIV protease inhibitors. We report here the development of a novel class of N1-aryl-propane-1,3-diamine compounds using a structure-based approach that selectively inhibit the activity of the bromodomain of the human transcriptional co-activator PCAF, of which association with the HIV trans-activator Tat is essential for transcription and replication of the integrated HIV provirus.
Organizational Affiliation:
Structural Biology Program, Department of Physiology and Biophysics, Mount Sinai School of Medicine, New York University, One Gustave L. Levy Place, New York, New York 10029-6574, USA.