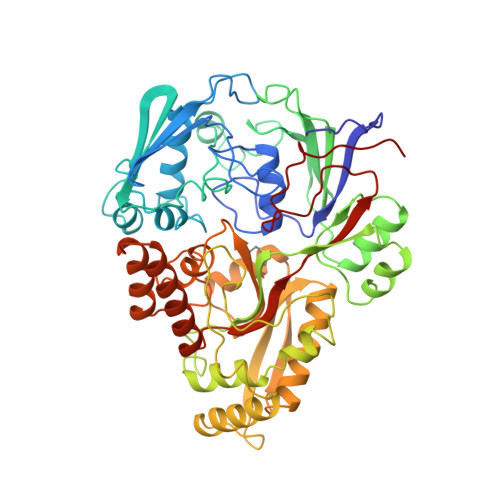
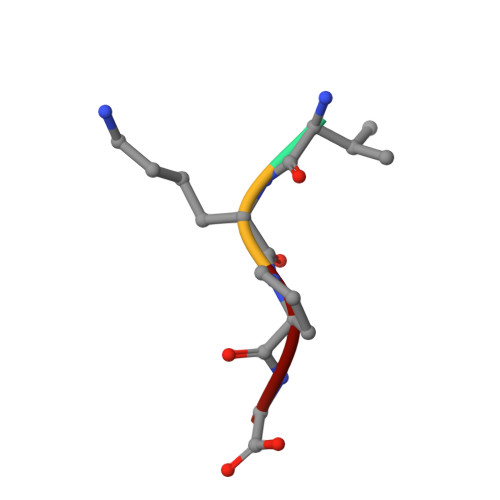
The structural basis of sequence-independent peptide binding by OppA protein.
Tame, J.R., Murshudov, G.N., Dodson, E.J., Neil, T.K., Dodson, G.G., Higgins, C.F., Wilkinson, A.J.(1994) Science 264: 1578-1581
- PubMed: 8202710
- DOI: https://doi.org/10.1126/science.8202710
- Primary Citation of Related Structures:
1OLA - PubMed Abstract:
Specific protein-ligand interactions are critical for cellular function, and most proteins select their partners with sharp discrimination. However, the oligopeptide-binding protein of Salmonella typhimurium (OppA) binds peptides of two to five amino acid residues without regard to sequence. The crystal structure of OppA reveals a three-domain organization, unlike other periplasmic binding proteins. In OppA-peptide complexes, the ligands are completely enclosed in the protein interior, a mode of binding that normally imposes tight specificity. The protein fulfills the hydrogen bonding and electrostatic potential of the ligand main chain and accommodates the peptide side chains in voluminous hydrated cavities.
Organizational Affiliation:
Department of Chemistry, University of York, UK.