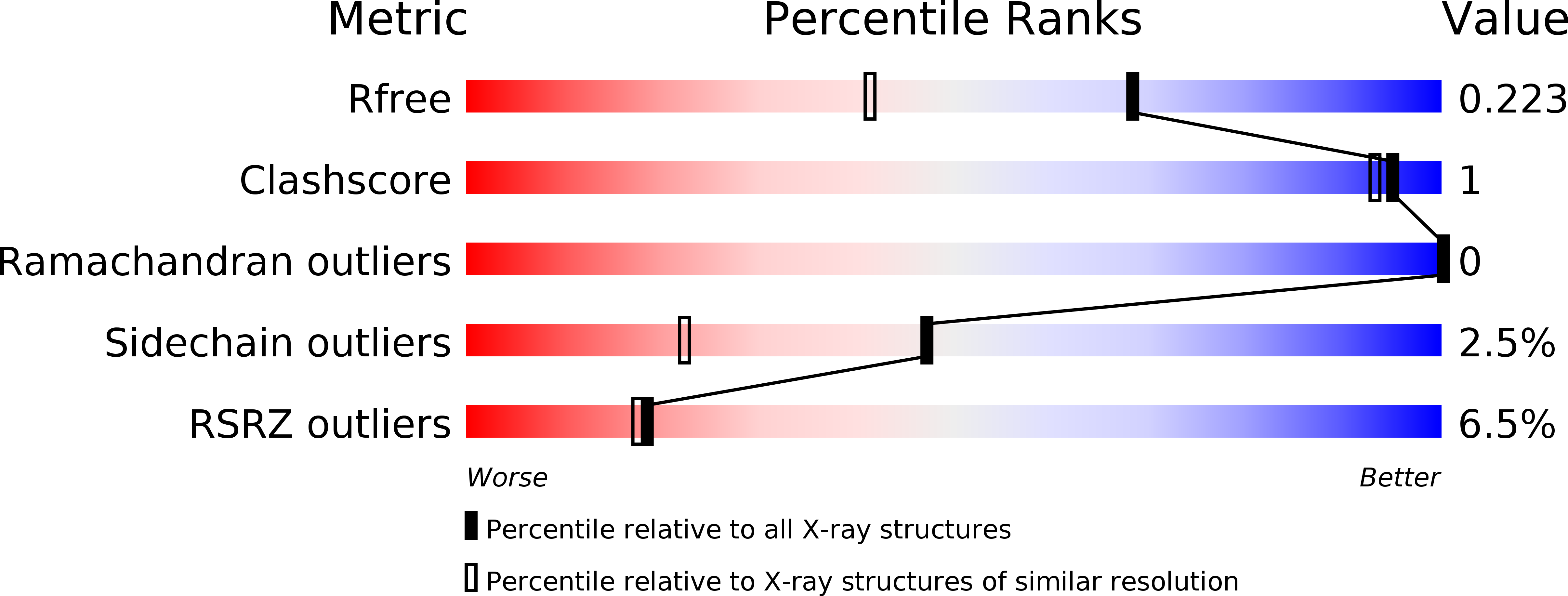
Dextranase from Penicillium Minioluteum. Reaction Course, Crystal Structure, and Product Complex
Larsson, A.M., Andersson, R., Stahlberg, J., Kenne, L., Jones, T.A.(2003) Structure 11: 1111
- PubMed: 12962629
- DOI: https://doi.org/10.1016/s0969-2126(03)00147-3
- Primary Citation of Related Structures:
1OGM, 1OGO - PubMed Abstract:
Dextranase catalyzes the hydrolysis of the alpha-1,6-glycosidic linkage in dextran polymers. The structure of dextranase, Dex49A, from Penicillium minioluteum was solved in the apo-enzyme and product-bound forms. The main domain of the enzyme is a right-handed parallel beta helix, which is connected to a beta sandwich domain at the N terminus. In the structure of the product complex, isomaltose was found to bind in a crevice on the surface of the enzyme. The glycosidic oxygen of the glucose unit in subsite +1 forms a hydrogen bond to the suggested catalytic acid, Asp395. By NMR spectroscopy the reaction course was shown to occur with net inversion at the anomeric carbon, implying a single displacement mechanism. Both Asp376 and Asp396 are suitably positioned to activate the water molecule that performs the nucleophilic attack. A new clan that links glycoside hydrolase families 28 and 49 is suggested.
Organizational Affiliation:
Department of Cell and Molecular Biology, Biomedical Centre, University of Uppsala, Box 596, SE-751 24 Uppsala, Sweden.