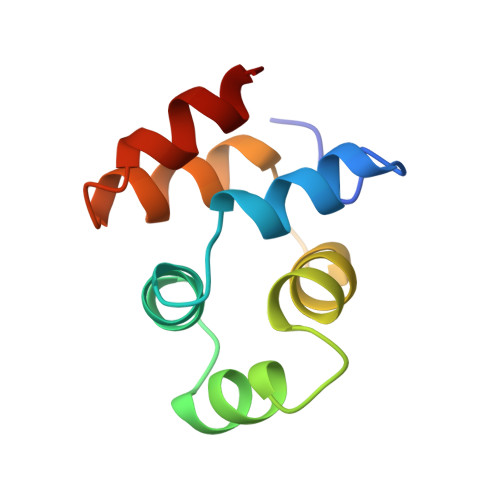
NMR structure of the death domain of the p75 neurotrophin receptor.
Liepinsh, E., Ilag, L.L., Otting, G., Ibanez, C.F.(1997) EMBO J 16: 4999-5005
- PubMed: 9305641
- DOI: https://doi.org/10.1093/emboj/16.16.4999
- Primary Citation of Related Structures:
1NGR - PubMed Abstract:
The intracellular domain of the p75 neurotrophin receptor (p75ICD) lacks catalytic activity but contains a motif similar to death domains found in the cytoplasmic regions of members of the tumor necrosis factor receptor family and their downstream targets. Although some aspects of the signaling pathways downstream of p75 have been elucidated recently, mechanisms of receptor activation and proximal signaling events are unknown. Here we report the nuclear magnetic resonance (NMR) structure of the 145 residue long p75ICD. The death domain of p75ICD consists of two perpendicular sets of three helices packed into a globular structure. The polypeptide segment connecting the transmembrane and death domains as well as the serine/threonine-rich C-terminal end are highly flexible in p75ICD. Unlike the death domains involved in signaling by the TNF receptor and Fas, p75ICD does not self-associate in solution. A surface area devoid of charged residues in the p75ICD death domain may indicate a potential site of interaction with downstream targets.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Karolinska Institute, Stockholm, Sweden.