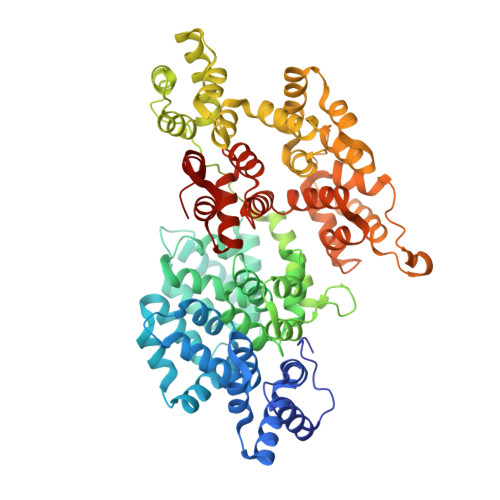
Structural and Dynamic Changes in Human Annexin VI Induced by a Phosphorylation-Mimicking Mutation, T356D
Freye-Minks, C., Kretsinger, R.H., Creutz, C.E.(2003) Biochemistry 42: 620-630
- PubMed: 12534274
- DOI: https://doi.org/10.1021/bi026742h
- Primary Citation of Related Structures:
1M9I - PubMed Abstract:
Phosphorylation of some members of the annexin family of proteins may play a significant role in controlling their calcium-dependent interactions with membranes. Recent electron microscopic studies of annexin VI revealed that the protein's two core domains exhibit a great degree of flexibility and are able to undergo a relative conformational change that could potentially initiate contacts between membranes [Avila-Sakar, A. J., et al. (2000) J. Struct. Biol. 130, 54-62]. To assess the possibility of a regulatory role of phosphorylation in this behavior, the crystal structure of a phosphorylation-mimicking mutant (T356D in the flexible connector region of human annexin VI) was determined to 2.65 A resolution. When the mutant is compared to the wild-type annexin VI, subtle differences are seen at the site of the mutation, while larger changes are evident in one of the calcium-binding loops and in the presence of five calcium ions. Furthermore, biochemical studies provide evidence for additional conformational differences between the T356D and wild-type solution structures. Fluorescence emission and acrylamide quenching suggest a higher level of solvent exposure of Trp-343 in the connector region of T356D in the presence of calcium. Comparisons of retardation coefficients in native gel electrophoresis reveal that T356D has a more extended shape, while proteolytic studies show a greater accessibility of a trypsin cleavage site inside the linker region, indicating a conformation more open than the wild-type form. These data provide insights into a possible regulatory mechanism leading to a higher degree of flexibility and possibly a higher calcium binding affinity of annexin VI upon phosphorylation.
Organizational Affiliation:
Department of Pharmacology, University of Virginia, Charlottesville, Virginia 22908, USA. cf9c@virginia.edu