NMR Determination of the Conformation of a Trimethylene Interstrand Cross-Link in an Oligodeoxynucleotide Duplex Containing a 5'-d(GpC) Motif
Dooley, P.D., Zhang, M., Korbel, G.A., Nechev, L.V., Harris, C.M., Stone, M.P., Harris, T.M.(2003) J Am Chem Soc 125: 62-72
- PubMed: 12515507
- DOI: https://doi.org/10.1021/ja0207798
- Primary Citation of Related Structures:
1LUH - PubMed Abstract:
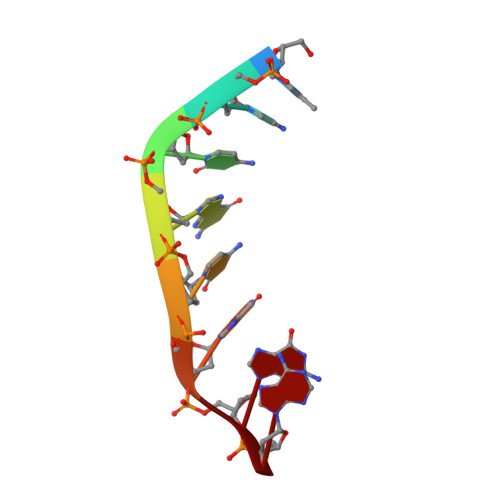
Malondialdehyde interstrand cross-links in DNA show strong preference for 5'-d(CpG) sequences. The cross-links are unstable and a trimethylene cross-link has been used as a surrogate for structural studies. A previous structural study of the 5'-d(CpG) cross-link in the sequence 5'-d(AGGCGCCT), where G is the modified nucleotide, by NMR spectroscopy and molecular dynamics using a simulated annealing protocol showed the guanine residues and the tether lay approximately in a plane such that the trimethylene tether and probably the malondialdehyde tether, as well, could be accommodated without major disruptions of duplex structure [Dooley et al. J. Am Chem. Soc. 2001, 123, 1730-1739]. The trimethylene cross-link has now been studied in a GpC motif using the reverse sequence. The structure lacks the planarity seen with the 5'-d(CpG) sequence and is skewed about the trimethylene cross-link. Melting studies indicate that the trimethylene cross-link is thermodynamically less stable in the GpC motif than in the 5-d(CpG). Furthermore, lack of planarity of the GpC cross-link precludes making an isosteric replacement of the trimethylene tether by malondialdehyde. A similar argument can be used to explain the 5'-d(CpG) preference for interchain cross-linking by acrolein.
Organizational Affiliation:
Department of Chemistry and Center in Molecular Toxicology, Vanderbilt University, Nashville, TN 37235, USA.