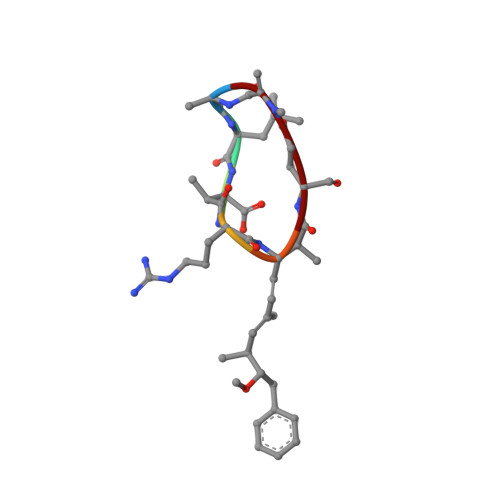
Conformational studies of microcystin-LR using NMR spectroscopy and molecular dynamics calculations.
Trogen, G.B., Annila, A., Eriksson, J., Kontteli, M., Meriluoto, J., Sethson, I., Zdunek, J., Edlund, U.(1996) Biochemistry 35: 3197-3205
- PubMed: 8605154
- DOI: https://doi.org/10.1021/bi952368s
- Primary Citation of Related Structures:
1LCM - PubMed Abstract:
NMR spectroscopy in aqueous and dimethyl sulfoxide/water solutions is used to determine the three-dimensional structures of microcystin-LR, a cyclic cyanobacterial heptapeptide toxin which is a potent inhibitor of type 1 and type 2A protein phosphatases. The conformations of this toxic peptide are studied using a simulated annealing (SA) protocol followed by refined SA calculations in vacuo and free MD simulations in water. Only one conformational family in each solvent is found. The peptide ring has a saddle-shaped form, essentially the same in both solvents. The structural difference observed between the two solution structures is located to the part consisting of Mdha, Ala, and Leu. This peptide segment is not present in nodularin, a cyclic pentapeptide of similar toxicity. The Arg side chain is very flexible, while the side chain of Leu is well defined. The side chain of Adda, essential for toxicity, is constrained in the vicinity of the backbone ring but appears to be flexible in the more remote part.
Organizational Affiliation:
Department of Organic Chemistry, Umea University, Sweden.