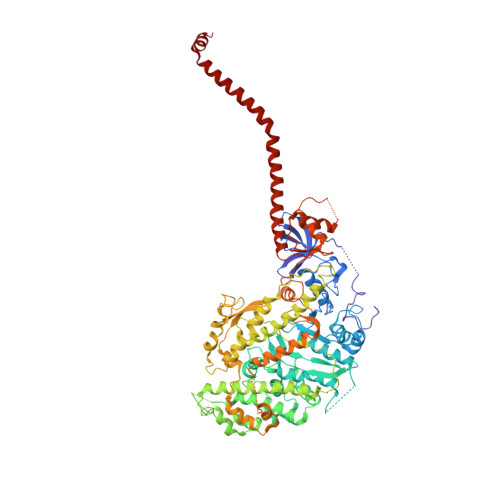
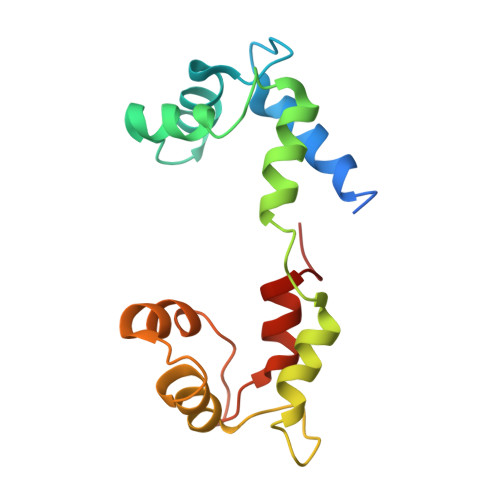
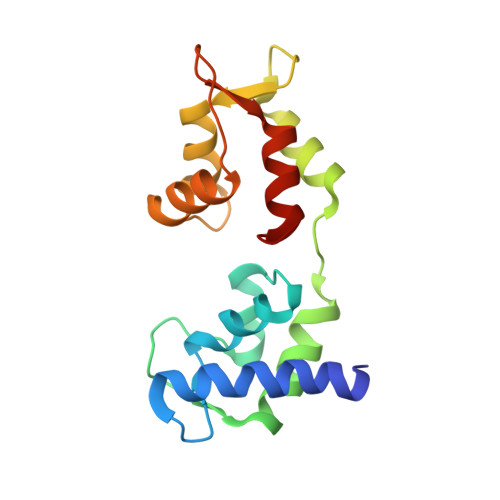
Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor.
Himmel, D.M., Gourinath, S., Reshetnikova, L., Shen, Y., Szent-Gyorgyi, A.G., Cohen, C.(2002) Proc Natl Acad Sci U S A 99: 12645-12650
- PubMed: 12297624
- DOI: https://doi.org/10.1073/pnas.202476799
- Primary Citation of Related Structures:
1KK7, 1KK8, 1KQM, 1KWO, 1L2O - PubMed Abstract:
Here we report a 2.3-A crystal structure of scallop myosin S1 complexed with ADP.BeF(x), as well as three additional structures (at 2.8-3.8 A resolution) for this S1 complexed with ATP analogs, some of which are cross-linked by para-phenyl dimaleimide, a short intramolecular cross-linker. In all cases, the complexes are characterized by an unwound SH1 helix first seen in an unusual 2.5-A scallop myosin-MgADP structure and described as corresponding to a previously unrecognized actin-detached internally uncoupled state. The unwinding of the SH1 helix effectively uncouples the converter/lever arm module from the motor and allows cross-linking by para-phenyl dimaleimide, which has been shown to occur only in weak actin-binding states of the molecule. Mutations near the metastable SH1 helix that disable the motor can be accounted for by viewing this structural element as a clutch controlling the transmission of torque to the lever arm. We have also determined a 3.2-A nucleotide-free structure of scallop myosin S1, which suggests that in the near-rigor state there are two conformations in the switch I loop, depending on whether nucleotide is present. Analysis of the subdomain motions in the weak actin-binding states revealed by x-ray crystallography, together with recent electron microscopic results, clarify the mechanical roles of the parts of the motor in the course of the contractile cycle and suggest how strong binding to actin triggers both the power stroke and product release.
Organizational Affiliation:
Rosenstiel Basic Medical Sciences Research Center, Waltham, MA 02454-9110, USA.