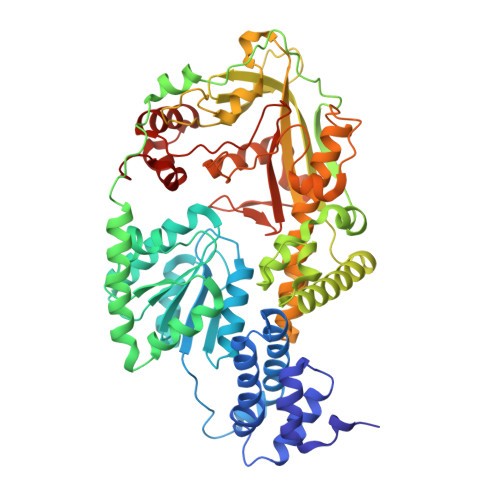
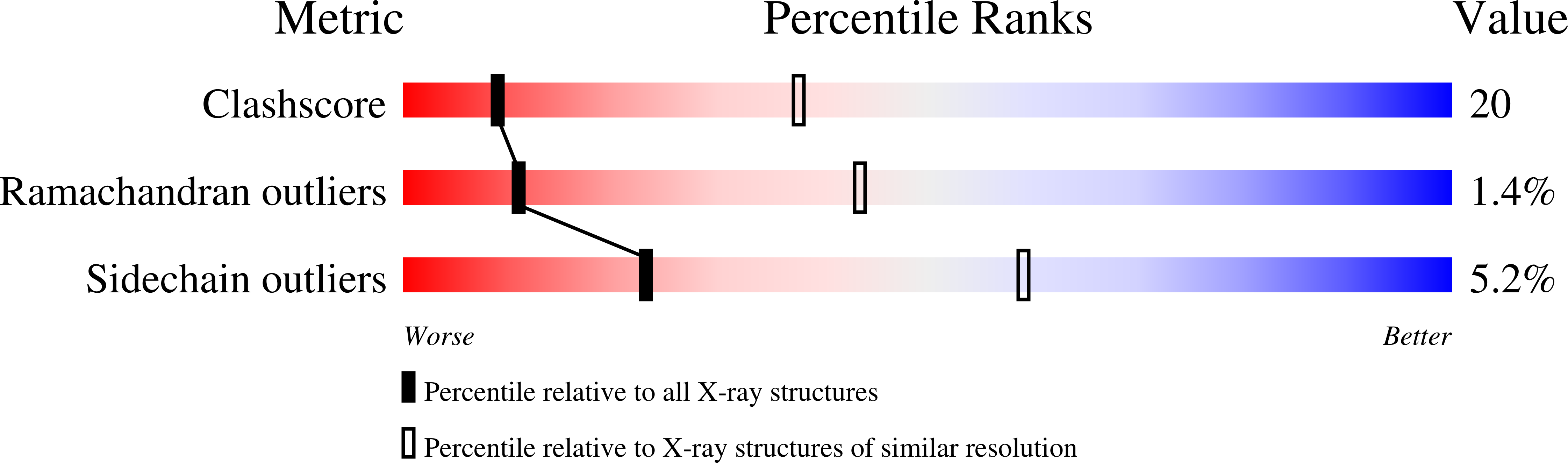Domain alternation switches B(12)-dependent methionine synthase to the activation conformation.
Bandarian, V., Pattridge, K.A., Lennon, B.W., Huddler, D.P., Matthews, R.G., Ludwig, M.L.(2002) Nat Struct Biol 9: 53-56
- PubMed: 11731805
- DOI: https://doi.org/10.1038/nsb738
- Primary Citation of Related Structures:
1K7Y, 1K98 - PubMed Abstract:
B(12)-dependent methionine synthase (MetH) from Escherichia coli is a large modular protein that uses bound cobalamin as an intermediate methyl carrier. Major domain rearrangements have been postulated to explain how cobalamin reacts with three different substrates: homocysteine, methyltetrahydrofolate and S-adenosylmethionine (AdoMet). Here we describe the 3.0 A structure of a 65 kDa C-terminal fragment of MetH that spans the cobalamin- and AdoMet-binding domains, arranged in a conformation suitable for the methyl transfer from AdoMet to cobalamin that occurs during activation. In the conversion to the activation conformation, a helical domain that capped the cofactor moves 26 A and rotates by 63 degrees, allowing formation of a new interface between cobalamin and the AdoMet-binding (activation) domain. Interactions with the MetH activation domain drive the cobalamin away from its binding domain in a way that requires dissociation of the axial cobalt ligand and, thereby, provide a mechanism for control of the distribution of enzyme conformations.
Organizational Affiliation:
Biophysics Research Division and Department of Biological Chemistry, University of Michigan, Ann Arbor, Michigan 48109-1055, USA.