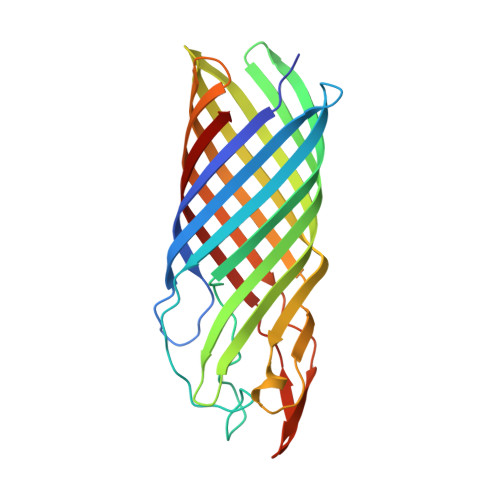
Crystal structure of the OpcA integral membrane adhesin from Neisseria meningitidis.
Prince, S.M., Achtman, M., Derrick, J.P.(2002) Proc Natl Acad Sci U S A 99: 3417-3421
- PubMed: 11891340
- DOI: https://doi.org/10.1073/pnas.062630899
- Primary Citation of Related Structures:
1K24 - PubMed Abstract:
OpcA is an integral outer membrane protein from Neisseria meningitidis, the causative agent of meningococcal meningitis and septicemia. It mediates the adhesion of N. meningitidis to epithelial and endothelial cells by binding to vitronectin and proteoglycan cell-surface receptors. Here, we report the determination of the crystal structure of OpcA to 2.0 A resolution. OpcA adopts a 10-stranded beta-barrel structure with extensive loop regions that protrude above the predicted surface of the membrane. The second external loop adopts an unusual conformation, traversing the axis of the beta-barrel and apparently blocking formation of a pore through the membrane. Loops 2, 3, 4, and 5 associate to form one side of a crevice in the external surface of the structure, the other side being formed by loop 1. The crevice is lined by positively charged residues and would form an ideal binding site for proteoglycan polysaccharide. The structure, therefore, suggests a model for how adhesion of this important human pathogen to proteoglycan is mediated at the molecular level.
Organizational Affiliation:
Department of Biomolecular Sciences, University of Manchester Institute of Science and Technology, Sackville Street, Manchester M60 1QD, United Kingdom.