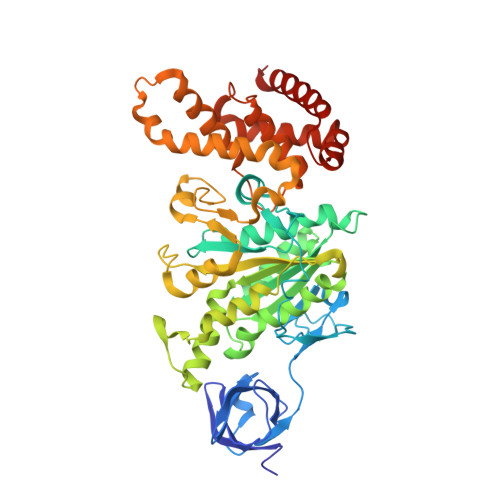
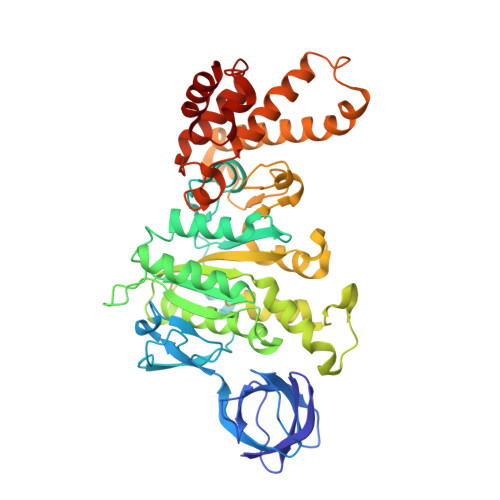
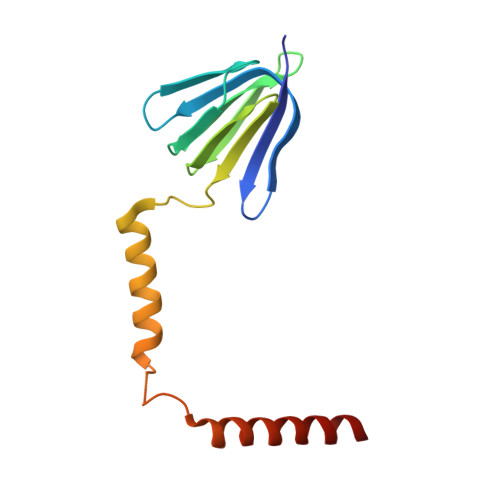
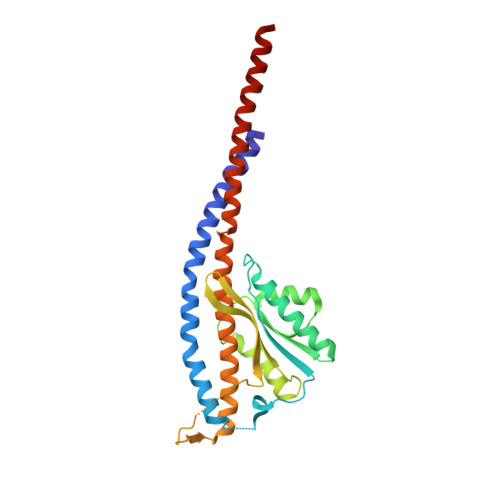
The conformation of the epsilon- and gamma-subunits within the Escherichia coli F(1) ATPase.
Hausrath, A.C., Capaldi, R.A., Matthews, B.W.(2001) J Biol Chem 276: 47227-47232
- PubMed: 11585832
- DOI: https://doi.org/10.1074/jbc.M107536200
- Primary Citation of Related Structures:
1JNV - PubMed Abstract:
F(1) is the water-soluble portion of the ubiquitous F(1)F(0) ATP synthase. Its structure includes three alpha- and three beta-subunits, arranged as a hexameric disc, plus a gamma-subunit that penetrates the center of the disc akin to an axle. Recently Hausrath et al. (Hausrath, A. C., Grüber, G., Matthews, B. W., and Capaldi, R. A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 13697-13702) obtained an electron density map of E. coli F(1) at 4.4-A resolution in which the coiled-coil alpha-helices of the gamma-subunit could be seen to extend 45 A from the base of the alpha(3)beta(3) hexamer. Subsequently the structure of a truncated form of the E. coli gamma-subunit in complex with epsilon has been described (Rodgers, A. J. W., and Wilce, M. C. J. (2000) Nat. Struct. Biol. 7, 1051-1054). In the present study the 4.4-A resolution electron density map of E. coli F(1) is re-evaluated in light of the newly available data on the gamma- and epsilon-subunits. It is shown that the map of the F(1) complex is consistent with the structure of the isolated subunits. When E. coli F(1) is compared with that from beef heart, the structures of the E. coli gamma- and epsilon-subunits are seen to be generally similar to their counterparts in the bovine enzyme but to undergo major shifts in position. In particular, the two long, coiled-coil alpha-helices that lie along the axis of F(1) both unwind and rotate. Also the epsilon-subunit rotates around the axis by 81 degrees and undergoes a net translation of about 23 A. It is argued that these large-scale changes in conformation reflect distinct functional states that occur during the rotation of the gamma-subunit within the alpha(3)beta(3) hexamer.
Organizational Affiliation:
Institute of Molecular Biology, Howard Hughes Medical Institute, University of Oregon, Eugene, OR 97403-1229, USA.