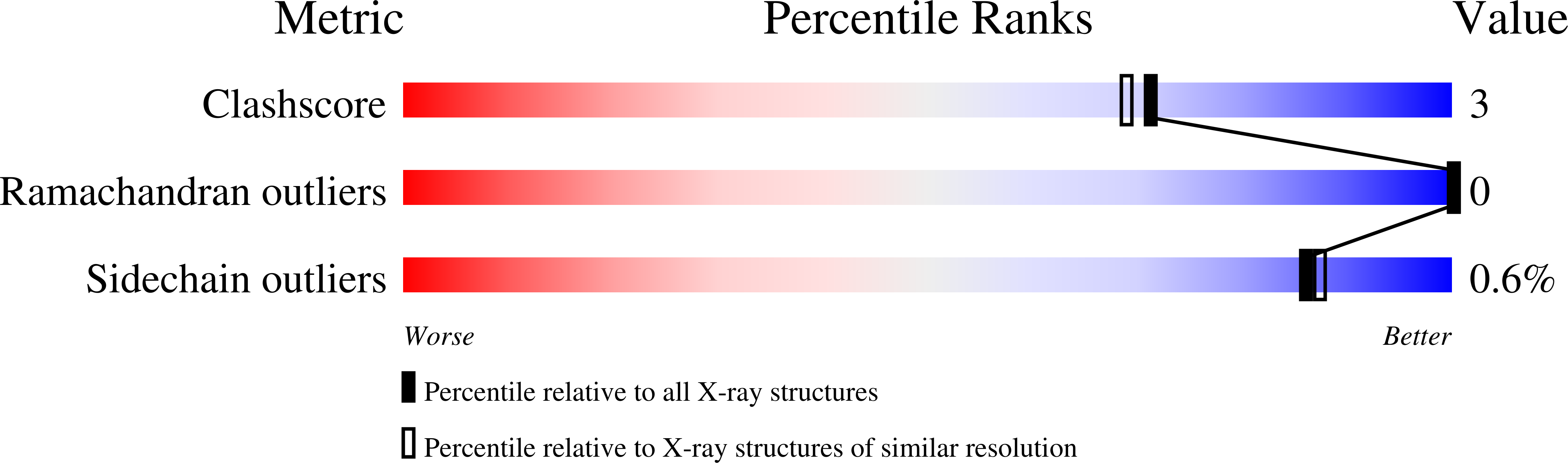Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms.
Notenboom, V., Boraston, A.B., Kilburn, D.G., Rose, D.R.(2001) Biochemistry 40: 6248-6256
- PubMed: 11371186
- DOI: https://doi.org/10.1021/bi0101704
- Primary Citation of Related Structures:
1I82, 1I8A, 1I8U - PubMed Abstract:
The C-terminal module of the thermostable Thermotoga maritima xylanase 10A (CBM9-2) is a family 9 carbohydrate-binding module that binds to amorphous and crystalline cellulose and a range of soluble di- and monosaccharides as well as to cello and xylo oligomers of different degrees of polymerization [Boraston, A. B., Creagh, A. L., Alam, Md. M., Kormos, J. M., Tomme, P., Haynes, C. A., Warren, R. A. J., and Kilburn, D. G. (2001) Biochemistry 40, 6240-6247]. The crystal structure of CBM9-2 has been determined by the multiwavelength anomalous dispersion method to 1.9 A resolution. CBM9-2 assumes a beta-sandwich fold and contains three metal binding sites. The bound metal atoms, which are most likely calcium cations, are in an octahedral coordination. The crystal structures of CBM9-2 in complex with glucose and cellobiose were also determined in order to identify the sugar-binding site and provide insight into the structural basis for sugar binding by CBM9-2. The sugar-binding site is a solvent-exposed slot sufficient in depth, width, and length to accommodate a disaccharide. Two tryptophan residues are stacked together on the surface of the protein forming the sugar-binding site. From the complex structures with glucose and cellobiose, it was inferred that CBM9-2 binds exclusively to the reducing end of mono-, di-, and oligosaccharides with an intricate hydrogen-bonding network involving mainly charged residues, as well as stacking interactions by Trp175 and Trp71. The binding interactions are limited to disaccharides as was expected from calorimetric data. Comparison of the glucose and cellobiose complexes revealed surprising differences in binding of these two substrates by CBM9-2. Cellobiose was found to bind in a distinct orientation from glucose, while still maintaining optimal stacking and electrostatic interactions with the reducing end sugar.
Organizational Affiliation:
Protein Engineering Network of Centres of Excellence, Ontario Cancer Institute, University of Toronto, Toronto, Ontario, Canada.