The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Benson, S.D., Bamford, J.K., Bamford, D.H., Burnett, R.M.(2002) Acta Crystallogr D Biol Crystallogr 58: 39-59
- PubMed: 11752778
- DOI: https://doi.org/10.1107/s0907444901017279
- Primary Citation of Related Structures:
1HQN, 1HX6 - PubMed Abstract:
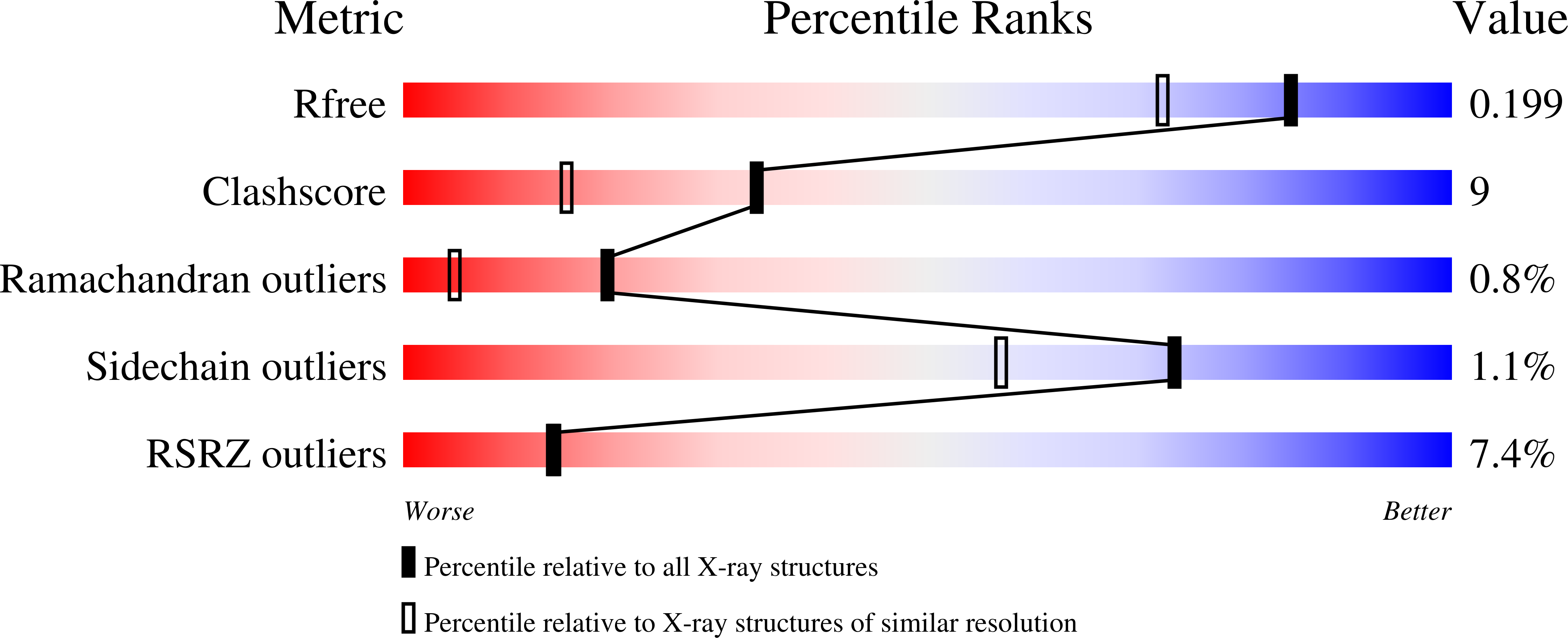
P3 has been imaged with X-ray crystallography to reveal a trimeric molecule with strikingly similar characteristics to hexon, the major coat protein of adenovirus. The structure of native P3 has now been extended to 1.65 A resolution (R(work) = 19.0% and R(free) = 20.8%). The new high-resolution model shows that P3 forms crystals through hydrophobic patches solvated by 2-methyl-2,4-pentanediol molecules. It reveals details of how the molecule's high stability may be achieved through ordered solvent in addition to intra- and intersubunit interactions. Of particular importance is a 'puddle' at the top of the molecule containing a four-layer deep hydration shell that cross-links a complex structural feature formed by 'trimerization loops'. These loops also link subunits by extending over a neighbor to reach the third subunit in the trimer. As each subunit has two eight-stranded viral jelly rolls, the trimer has a pseudo-hexagonal shape to allow close packing in its 240 hexavalent capsid positions. Flexible regions in P3 facilitate these interactions within the capsid and with the underlying membrane. A selenometh-ionine P3 derivative, with which the structure was solved, has been refined to 2.2 A resolution (R(work) = 20.1% and R(free) = 22.8%). The derivatized molecule is essentially unchanged, although synchrotron radiation has the curious effect of causing it to rotate about its threefold axis. P3 is a second example of a trimeric 'double-barrel' protein that forms a stable building block with optimal shape for constructing a large icosahedral viral capsid. A major difference is that hexon has long variable loops that distinguish different adenovirus species. The short loops in P3 and the severe constraints of its various interactions explain why the PRD1 family has highly conserved coat proteins.
Organizational Affiliation:
The Wistar Institute, 3601 Spruce Street, Philadelphia, PA 19104, USA.