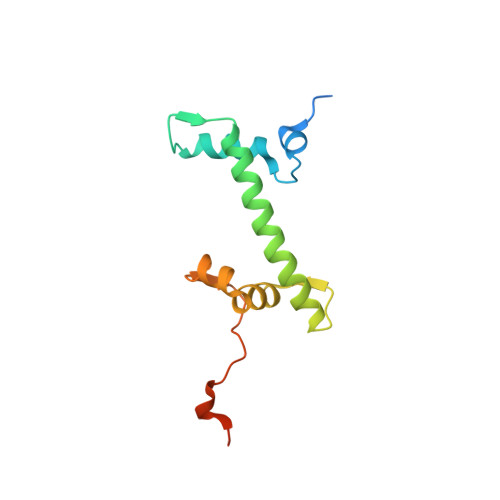
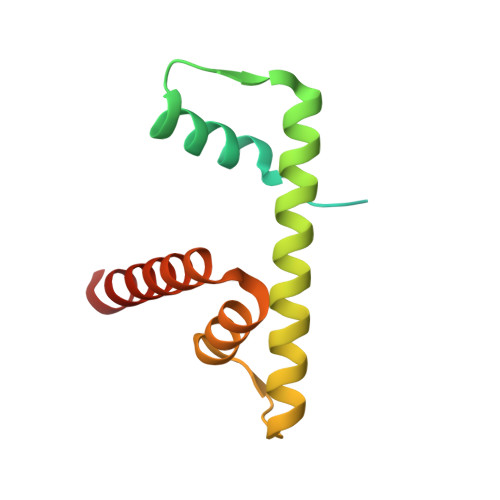
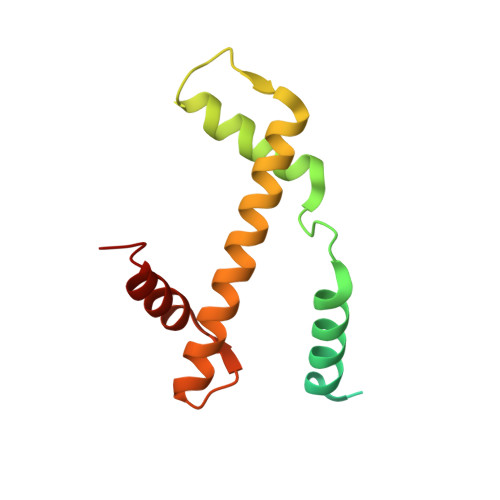
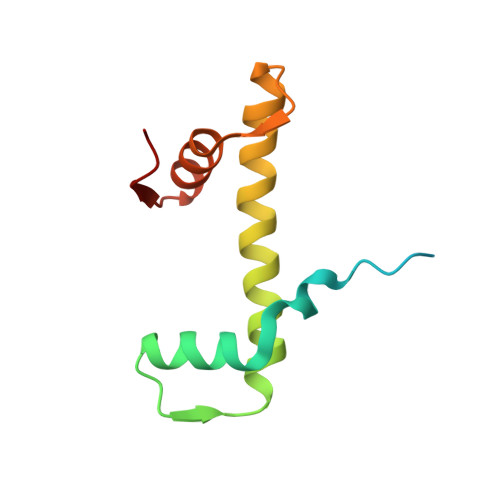
Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Chantalat, L., Nicholson, J.M., Lambert, S.J., Reid, A.J., Donovan, M.J., Reynolds, C.D., Wood, C.M., Baldwin, J.P.(2003) Acta Crystallogr D Biol Crystallogr 59: 1395-1407
- PubMed: 12876341
- DOI: https://doi.org/10.1107/s0907444903011880
- Primary Citation of Related Structures:
1HQ3 - PubMed Abstract:
The structure of the native chicken histone octamer, crystallized in 2 M KCl, 1.35 M potassium phosphate pH 6.9, has been refined at 2.15 A resolution to a final R factor of 21.4% and an R(free) of 25.2%. Unique crystal-packing interactions between histone-core octamers are strong and one of them (area 4000 A(2)) involves two chloride ions and direct interactions between six acidic amino-acid residues on one octamer and the equivalent number of basic residues on the next. These interactions are on the structured part of the octamer (not involving tails). Five phosphate ions, 23 chloride ions and 437 water molecules have been identified in the structure. The phosphate and some chloride ions bind to basic amino-acid residues that interact with DNA in the nucleosome. The binding of most of the anions and the packing interactions are unique to these crystals. In other respects, and including the positions of four chloride ions, the octamer structure is very close to that of octamers in nucleosome-core particle crystals, particularly with respect to 'docking' sequences of the histone H2As and H4s. These sequences together with the H2B-H4 four-helix bundles stabilize the histone structure in the nucleosome and prevent the dissociation of the (H2A-H2B) dimers from the (H3-H4)(2) tetramer. Possible reasons why this happens at high salt in the absence of DNA are given.
Organizational Affiliation:
Structural Biology, Galderma RandD, 635 Route des Lucioles, BP 87F-06902 Sophia Antipolis CEDEX, France.