X-ray structure of full-length annexin 1 and implications for membrane aggregation.
Rosengarth, A., Gerke, V., Luecke, H.(2001) J Mol Biol 306: 489-498
- PubMed: 11178908
- DOI: https://doi.org/10.1006/jmbi.2000.4423
- Primary Citation of Related Structures:
1HM6 - PubMed Abstract:
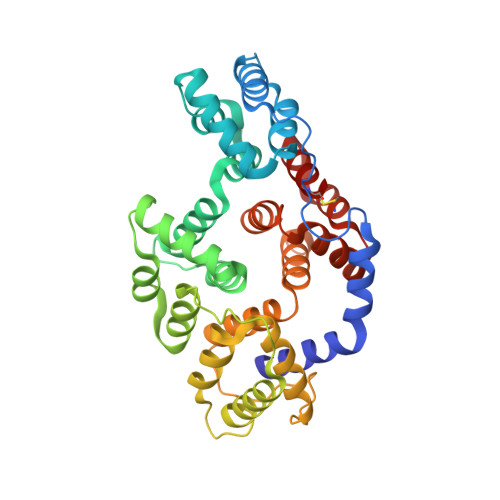
Annexins comprise a multigene family of Ca2+ and phospholipid- binding proteins. They consist of a conserved C-terminal or core domain that confers Ca2+-dependent phospholipid binding and an N-terminal domain that is variable in sequence and length and responsible for the specific properties of each annexin. Crystal structures of various annexin core domains have revealed a high degree of similarity. From these and other studies it is evident that the core domain harbors the calcium-binding sites that interact with the phospholipid headgroups. However, no structure has been reported of an annexin with a complete N-terminal domain. We have now solved the crystal structure of such a full-length annexin, annexin 1. Annexin 1 is active in membrane aggregation and its refined 1.8 A structure shows an alpha-helical N-terminal domain connected to the core domain by a flexible linker. It is surprising that the two alpha-helices present in the N-terminal domain of 41 residues interact intimately with the core domain, with the amphipathic helix 2-12 of the N-terminal domain replacing helix D of repeat III of the core. In turn, helix D is unwound into a flap now partially covering the N-terminal helix. Implications for membrane aggregation will be discussed and a model of aggregation based on the structure will be presented.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry and UCI Program in Macromolecular Structure, University of California, 3205 Biological Sciences II, Irvine, CA, 92697-3900, USA.