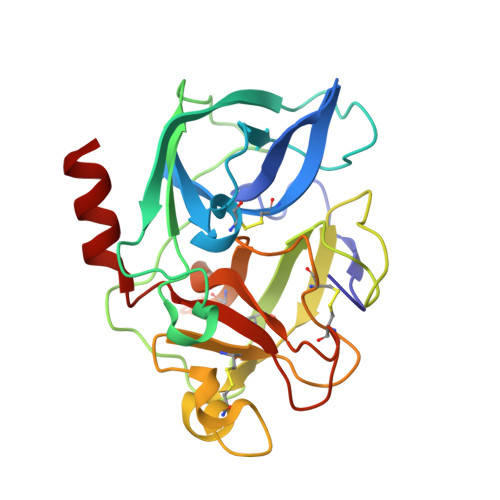
Kinetic and Crystallographic Analysis of Complexes Formed between Elastase and Peptides from Beta-Casein
Wright, P.A., Wilmouth, R.C., Clifton, I.J., Schofield, C.J.(2001) Eur J Biochem 268: 2969
- PubMed: 11358514
- DOI: https://doi.org/10.1046/j.1432-1327.2001.02186.x
- Primary Citation of Related Structures:
1H9L - PubMed Abstract:
Human beta-casomorphin-7 (NH2-Tyr-Pro-Phe-Val-Glu-Pro-Ile-CO2H) is a naturally occurring peptide inhibitor of elastase that has been shown to form an acyl-enzyme complex stable enough for X-ray crystallographic analysis at pH 5. To investigate the importance of the N-terminal residues of the beta-casomorphin-7 peptide for the inhibition of elastase, kinetic and crystallographic analyses were undertaken to identify the minimum number of residues required for effective formation of a stable complex between truncated beta-casomorphin-7 peptides and porcine pancreatic elastase (PPE). The results clearly demonstrate that significant inhibition of PPE can be effected by simple tri-, tetra-and pentapeptides terminating in a carboxylic acid. These results also suggest that in vivo regulation of protease activity could be mediated via short peptides as well as by proteins. Crystallographic analysis of the complex formed between N-acetyl-Val-Glu-Pro-Ile-CO2H and PPE at pH 5 (to 1.67 A resolution) revealed an active site water molecule in an analogous position to that observed in the PPE/beta-casomorphin-7 structure supportive of its assignment as the 'hydrolytic water' in the deacylation step of serine protease catalysis.
Organizational Affiliation:
The Dyson Perrins Laboratory and The Oxford Centre for Molecular Sciences, Oxford, UK.