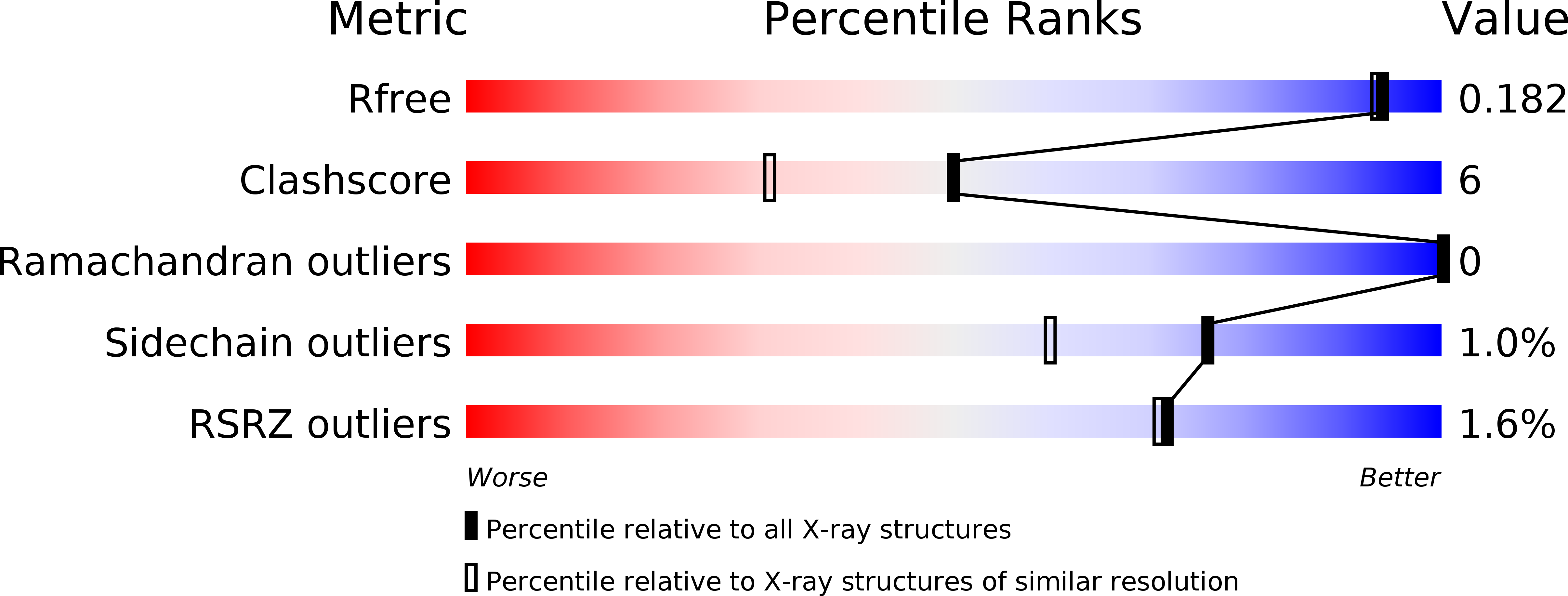
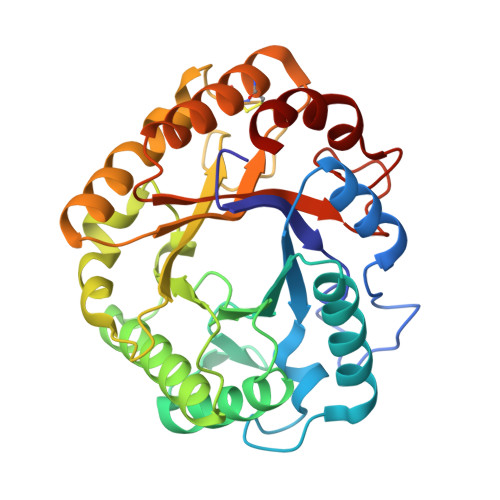
The 1.62 A Structure of Thermoascus Aurantiacus Endoglucanase: Completing the Structural Picture of Subfamilies in Glycoside Hydrolase Family 5
Lo Leggio, L., Larsen, S.(2002) FEBS Lett 523: 103-108
- PubMed: 12123813
- DOI: https://doi.org/10.1016/s0014-5793(02)02954-x
- Primary Citation of Related Structures:
1GZJ - PubMed Abstract:
The crystal structure of Thermoascus aurantiacus endoglucanase (Cel5A), a family 5 glycoside hydrolase, has been determined to 1.62 A resolution by multiple isomorphous replacement with anomalous scattering. It is the first report of a structure in the subfamily to which Cel5A belongs. Cel5A consists solely of a catalytic module with compact eight-fold beta/alpha barrel architecture. The length of the tryptophan-rich substrate binding groove suggests the presence of substrate binding subsites -4 to +3. Structural comparison shows that two glycines are completely conserved in the family, in addition to the two catalytic glutamates and six other conserved residues previously identified. Gly 44 in particular is part of a type IV C-terminal helix capping motif, whose disruption is likely to affect the position of an essential conserved arginine. One aromatic residue (Trp 170 in Cel5A), not conserved in term of sequence, is nonetheless spatially conserved in the substrate binding groove. Its role might be to force the bend that occurs in the polysaccharide chain on binding, thus favoring substrate distortion at subsite -1.
Organizational Affiliation:
Centre for Crystallographic Studies, Department of Chemistry, Chemical Institute, University of Copenhagen, Universitetsparken 5, DK-2100, Copenhagen, Denmark. leila@ccs.ki.ku.dk