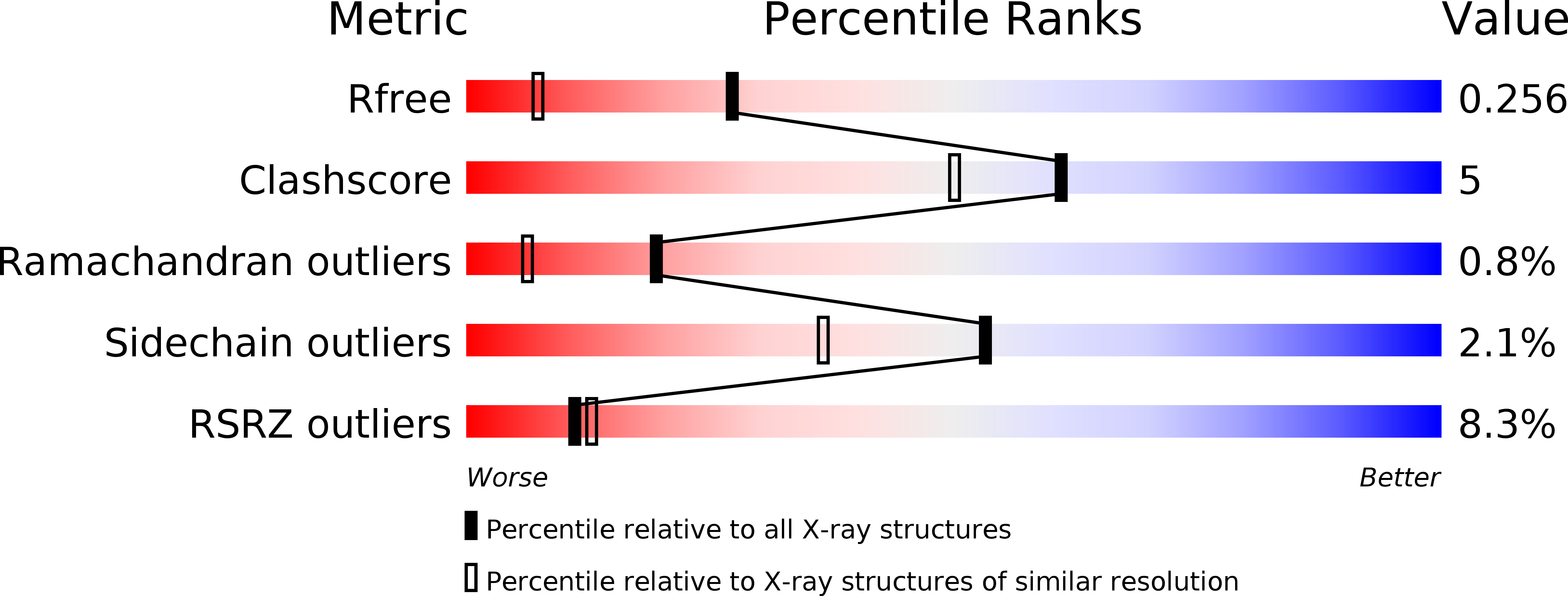
The Crystal Structure of Hpce, a Bifunctional Decarboxylase/Isomerase with a Multifunctional Fold.
Tame, J.R.H., Namba, K., Dodson, E.J., Roper, D.I.(2002) Biochemistry 41: 2982
- PubMed: 11863436
- DOI: https://doi.org/10.1021/bi015717t
- Primary Citation of Related Structures:
1GTT - PubMed Abstract:
The structure of the bifunctional enzyme HpcE (OPET decarboxylase/HHDD isomerase) from Escherichia coli shows that the protein consists of highly similar N and C terminal halves. Sequence matches suggest that this fold is widespread among different species, including man. Many of these homologues are uncharacterized but apparently connected with the metabolism of aromatic compounds. The domain shows similar topology to the C terminal domain of fumarylacetoacetate hydrolase (FAH), a functionally related enzyme, despite lacking significant overall sequence similarity. HpcE is known to catalyze two rather different reactions, and comparisons with FAH allow some tentative conclusions to be drawn about the active sites. Key mutations within the active site apparently allow enzymes with this fold to carry out a variety chemical processes.
Organizational Affiliation:
Protein Design Laboratory, Yokohama City University, Suehiro 1-7-29, Kanagawa 230-0045, Japan. jtame@tsurumi.yokohama-cu.ac.jp