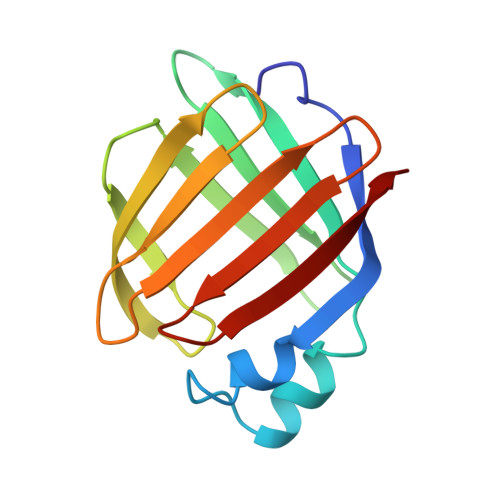
Three-dimensional structure of the muscle fatty-acid-binding protein isolated from the desert locust Schistocerca gregaria.
Haunerland, N.H., Jacobson, B.L., Wesenberg, G., Rayment, I., Holden, H.M.(1994) Biochemistry 33: 12378-12385
- PubMed: 7918460
- DOI: https://doi.org/10.1021/bi00207a004
- Primary Citation of Related Structures:
1FTP - PubMed Abstract:
The three-dimensional structure of the fatty-acid-binding protein isolated from the flight muscle of the desert locust Schistocerca gregaria has been solved and refined to a crystallographic R-value of 18.5% for all measured X-ray data from 30.0- to 2.2-A resolution. Crystals employed in the investigation were grown from 2.6 to 2.8 M ammonium sulfate solutions, buffered at pH 7.5 and containing 2-5% 2-methyl-2,4-pentanediol. They belonged to the space group P2(1) with unit cell dimensions of a = 61.6 A, b = 44.8 A, c = 63.9 A, and beta = 113.6 degrees and two molecules per asymmetric unit. The protein fold consists of ten strands of antiparallel beta-pleated sheet that wrap around to form a beta-barrel. In addition, there are two small alpha-helices and six type I, two type II, and two type II' turns. The two molecules pack in the asymmetric unit as a dimer with a local 2-fold rotational axis. The subunit-subunit interface involves amino acid side chains located in the area of the helix-turn-helix motif and the turn between beta-strands E and F. It is this area that has been speculated to form the portal through which fatty acids enter the binding cavity. There are 23 solvent molecules that are conserved between the two independent molecules in the asymmetric unit. Nine of these waters play important structural roles. A three-dimensional comparison between the insect and human muscle fatty-acid-binding proteins shows that their alpha-carbons superimpose with a root-mean-square deviation of 0.77 A for 89 structurally equivalent atoms.(ABSTRACT TRUNCATED AT 250 WORDS)
Organizational Affiliation:
Department of Biological Sciences, Simon Fraser University, Burnaby, Canada.