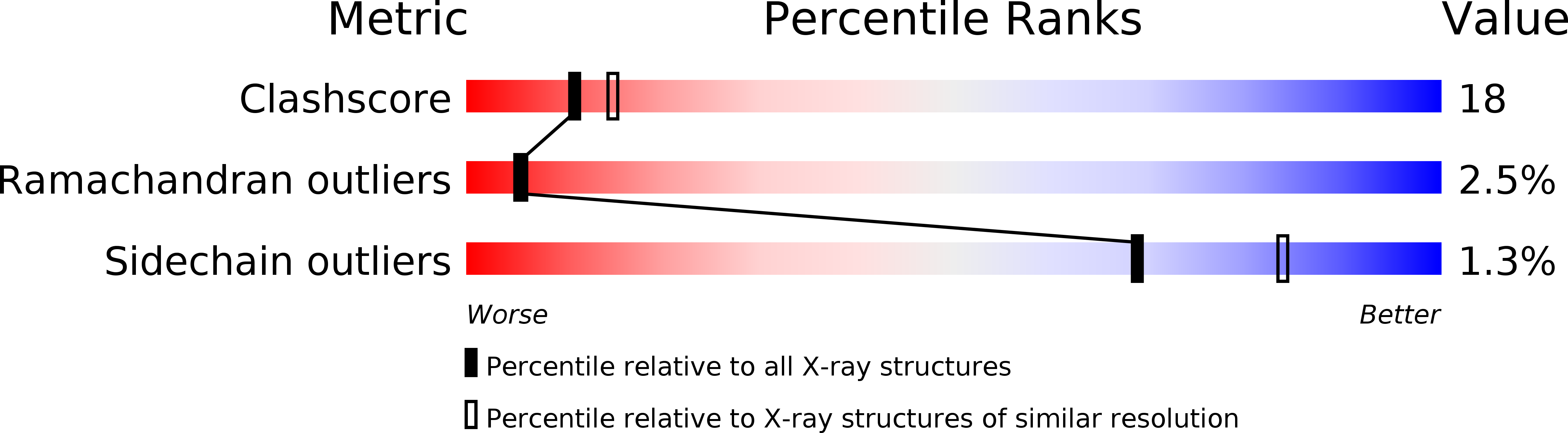Crystal structure of the human high-affinity IgE receptor.
Garman, S.C., Kinet, J.P., Jardetzky, T.S.(1998) Cell 95: 951-961
- PubMed: 9875849
- DOI: https://doi.org/10.1016/s0092-8674(00)81719-5
- Primary Citation of Related Structures:
1F2Q - PubMed Abstract:
Allergic responses result from the activation of mast cells by the human high-affinity IgE receptor. IgE-mediated allergic reactions may develop to a variety of environmental compounds, but the initiation of a response requires the binding of IgE to its high-affinity receptor. We have solved the X-ray crystal structure of the antibody-binding domains of the human IgE receptor at 2.4 A resolution. The structure reveals a highly bent arrangement of immunoglobulin domains that form an extended convex surface of interaction with IgE. A prominent loop that confers specificity for IgE molecules extends from the receptor surface near an unusual arrangement of four exposed tryptophans. The crystal structure of the IgE receptor provides a foundation for the development of new therapeutic approaches to allergy treatment.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Cell Biology, Northwestern University, Evanston, Illinois 60208, USA.