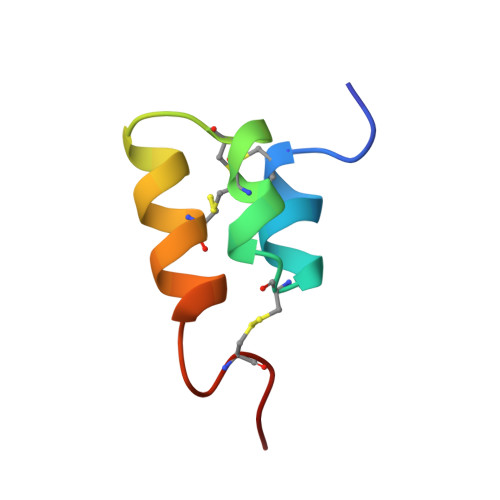
The NMR solution structure of the pheromone Er-2 from the ciliated protozoan Euplotes raikovi.
Ottiger, M., Szyperski, T., Luginbuhl, P., Ortenzi, C., Luporini, P., Bradshaw, R.A., Wuthrich, K.(1994) Protein Sci 3: 1515-1526
- PubMed: 7833811
- DOI: https://doi.org/10.1002/pro.5560030917
- Primary Citation of Related Structures:
1ERD - PubMed Abstract:
The NMR structure of the pheromone Er-2 from the ciliated protozoan Euplotes raikovi has been determined in aqueous solution. The structure of this 40-residue protein was calculated with the distance geometry program DIANA from 621 distance constraints and 89 dihedral angle constraints; the program OPAL was employed for the energy minimization. For a group of 20 conformers used to characterize the solution structure, the average pairwise RMS deviation from the mean structure calculated for the backbone heavy atoms N, C alpha, and C' of residues 3-37 was 0.31 A. The molecular architecture is dominated by an up-down-up bundle of 3 short helices of residues 5-11, 14-20, and 23-33, which is similar to the structures of the homologous pheromones Er-1 and Er-10. Novel structural features include a well-defined N-cap on the first helix, a 1-residue deletion in the second helix resulting in the formation of a 3(10)-helix rather than an alpha-helix as found in Er-1 and Er-10, and the simultaneous presence of 2 different conformations for the C-terminal tetrapeptide segment, i.e., a major conformation with the Leu 39-Pro 40 peptide bond in the trans form and a minor conformation with this peptide bond in the cis form.
Organizational Affiliation:
Institut für Molekularbiologie und Biophysik, Eidgenössische Technische Hochschule, Zürich, Switzerland.