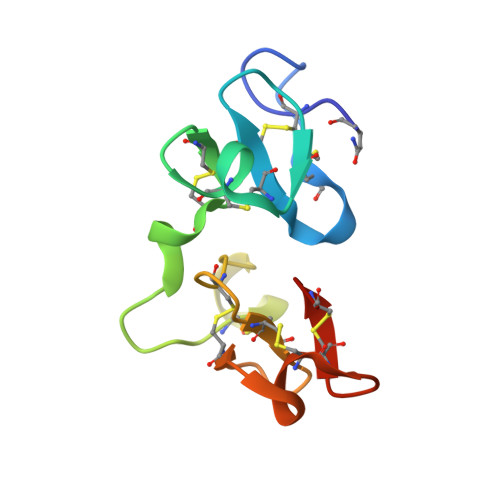
Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Saul, F.A., Rovira, P., Boulot, G., Damme, E.J., Peumans, W.J., Truffa-Bachi, P., Bentley, G.A.(2000) Structure 8: 593-603
- PubMed: 10873861
- DOI: https://doi.org/10.1016/s0969-2126(00)00142-8
- Primary Citation of Related Structures:
1EIS, 1EN2, 1ENM - PubMed Abstract:
Urtica dioica agglutinin (UDA), a monomeric lectin extracted from stinging nettle rhizomes, is specific for saccharides containing N-acetylglucosamine (GlcNAc). The lectin behaves as a superantigen for murine T cells, inducing the exclusive proliferation of Vbeta8.3(+) lymphocytes. UDA is unique among known T cell superantigens because it can be presented by major histocompatibility complex (MHC) molecules of both class I and II.
Organizational Affiliation:
Unité d'Immunologie Structurale (CNRS URA 2185), Institut Pasteur, Paris, France.