Crystal structure of SULT2A3, human hydroxysteroid sulfotransferase.
Pedersen, L.C., Petrotchenko, E.V., Negishi, M.(2000) FEBS Lett 475: 61-64
- PubMed: 10854859
- DOI: https://doi.org/10.1016/s0014-5793(00)01479-4
- Primary Citation of Related Structures:
1EFH - PubMed Abstract:
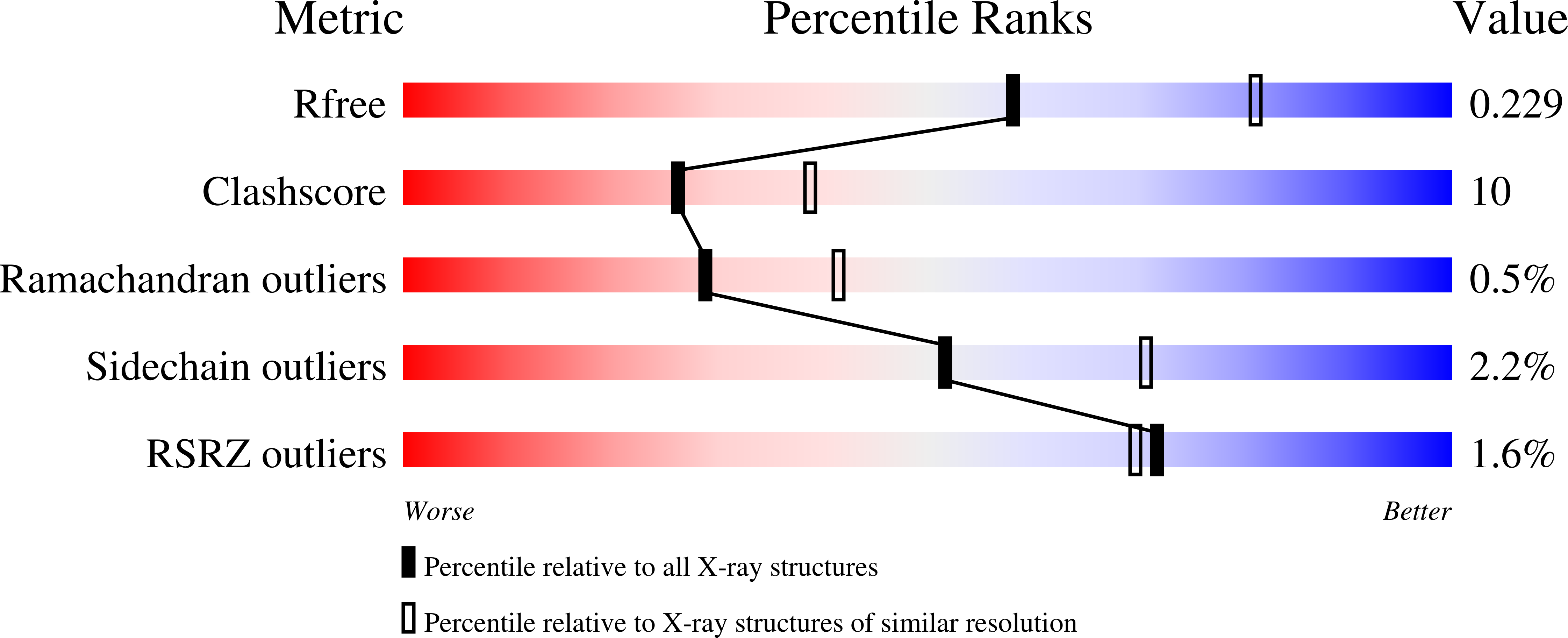
The crystal structure of SULT2A3 human hydroxysteroid sulfotransferase has been solved at 2.4 A resolution in the presence of 3'-phosphoadenosine 5'-phosphate (PAP). The overall structure is similar to those of SULT1 enzymes such as estrogen sulfotransferase and the PAP binding site is conserved, however, significant differences exist in the positions of loops Pro14-Ser20, Glu79-Ile82 and Tyr234-Gln244 in the substrate binding pocket. Moreover, protein interaction in the crystal structure has revealed a possible dimer-directed conformational alteration that may regulate the SULT activity.
Organizational Affiliation:
Pharmacogenetics Section, Laboratory of Reproductive and Developmental Toxicology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.