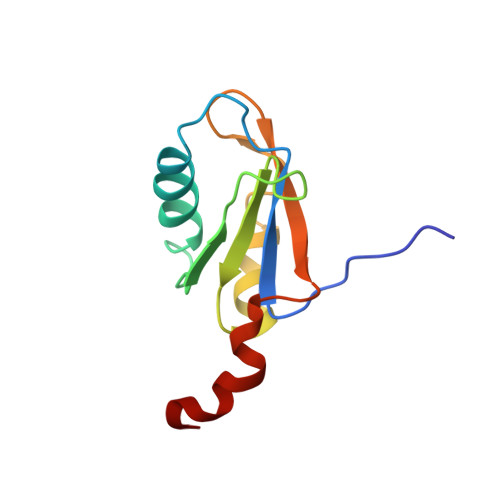
The NMR Structure of the 38kDa U1A Protein-Pie RNA Complex Reveals the Basis of Cooperativity in Regulation of Polyadenylation by Human U1A Protein
Varani, L., Gunderson, S.I., Mattaj, I.W., Kay, L.E., Neuhaus, D., Varani, G.(2000) Nat Struct Biol 7: 329
- PubMed: 10742179
- DOI: https://doi.org/10.1038/74101
- Primary Citation of Related Structures:
1DZ5 - PubMed Abstract:
The status of the poly(A) tail at the 3'-end of mRNAs controls the expression of numerous genes in response to developmental and extracellular signals. Poly(A) tail regulation requires cooperative binding of two human U1A proteins to an RNA regulatory region called the polyadenylation inhibition element (PIE). When bound to PIE RNA, U1A proteins also bind to the enzyme responsible for formation of the mature 3'-end of most eukaryotic mRNAs, poly(A) polymerase (PAP). The NMR structure of the 38 kDa complex formed between two U1A molecules and PIE RNA shows that binding cooperativity depends on helix C located at the end of the RNA-binding domain and just adjacent to the PAP-interacting domain of U1A. Since helix C undergoes a conformational change upon RNA binding, the structure shows that binding cooperativity and interactions with PAP occur only when U1A is bound to its cognate RNA. This mechanism ensures that the activity of PAP enzyme, which is essential to the cell, is only down regulated when U1A is bound to the U1A mRNA.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK.