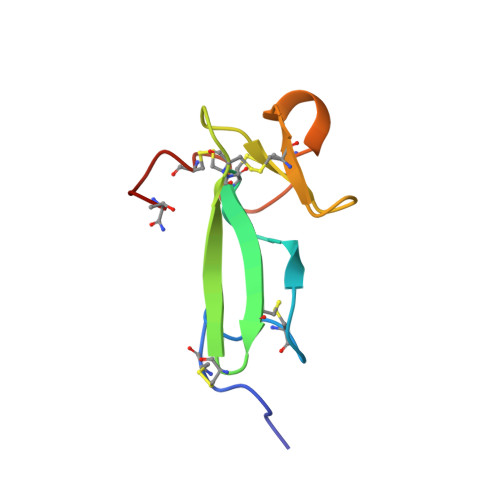
Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif.
Suetake, T., Tsuda, S., Kawabata, S., Miura, K., Iwanaga, S., Hikichi, K., Nitta, K., Kawano, K.(2000) J Biol Chem 275: 17929-17932
- PubMed: 10770921
- DOI: https://doi.org/10.1074/jbc.C000184200
- Primary Citation of Related Structures:
1DQC - PubMed Abstract:
Tachycitin, a 73-residue polypeptide having antimicrobial activity is present in the hemocyte of horseshoe crab (Tachypleus tridentatus). The first three-dimensional structure of invertebrate chitin-binding protein was determined for tachycitin using two-dimensional nuclear magnetic resonance spectroscopy. The measurements indicate that the structure of tachycitin is largely divided into N- and C-terminal domains; the former comprises a three-stranded beta-sheet and the latter a two-stranded beta-sheet following a short helical turn. The latter structural motif shares a significant tertiary structural similarity with the chitin-binding domain of plant chitin-binding protein. This result is thought to provide faithful experimental evidence to the recent hypothesis that chitin-binding proteins of invertebrates and plants are correlated by a convergent evolution process.
Organizational Affiliation:
Division of Biological Sciences, Graduate School of Science, Hokkaido University, Sapporo 060-0810, Japan. tsuda@hniri.go.jp