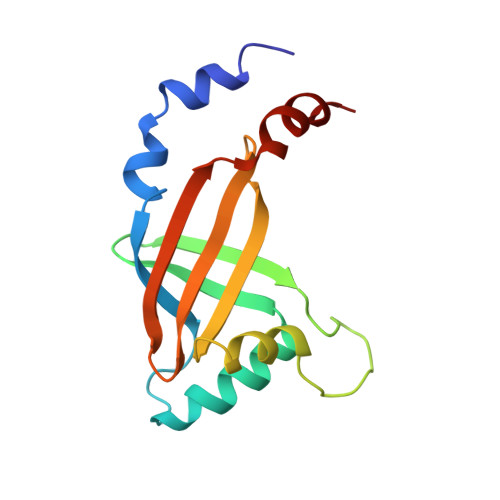
Molecular insights into PEBP2/CBF beta-SMMHC associated acute leukemia revealed from the structure of PEBP2/CBF beta
Goger, M., Gupta, V., Kim, W.Y., Shigesada, K., Ito, Y., Werner, M.H.(1999) Nat Struct Biol 6: 620-623
- PubMed: 10404215
- DOI: https://doi.org/10.1038/10664
- Primary Citation of Related Structures:
1CL3 - PubMed Abstract:
PEBP2/CBF is a heterodimeric transcription factor essential for genetic regulation of hematopoiesis and osteogenesis. DNA binding by PEBP2/CBF alpha is accomplished by a highly conserved DNA binding domain, the Runt domain (RD), whose structure adopts an S-type immunoglobulin fold when bound to DNA. The supplementary subunit beta enhances DNA binding by the RD in vitro, but its role in the control of gene expression has remained largely unknown in vivo. Chromosome 16 inversion creates a chimeric gene product fusing PEBP2/CBF beta to a portion of the smooth muscle myosin heavy chain (PEBP2/CBF beta-SMMHC) that is causally associated with the onset of acute myeloid leukemia in humans. The three-dimensional structure of PEBP2/CBF beta has been determined in solution and is shown to adopt a fold related to the beta-barrel oligomer binding motif. Direct analysis of a 43.6 kD ternary RD-beta-DNA complex identifies the likely surface of beta in contact with the RD. The structure of PEBP2/CBF beta enables a molecular understanding of the capacity of PEBP2/CBF beta-SMMHC to sequester PEBP2/CBF alpha in the cytoplasm and therefore provides a molecular basis for understanding leukemogenic transformation.
Organizational Affiliation:
Laboratory of Molecular Biophysics, The Rockefeller University, New York, NY 10021, USA.