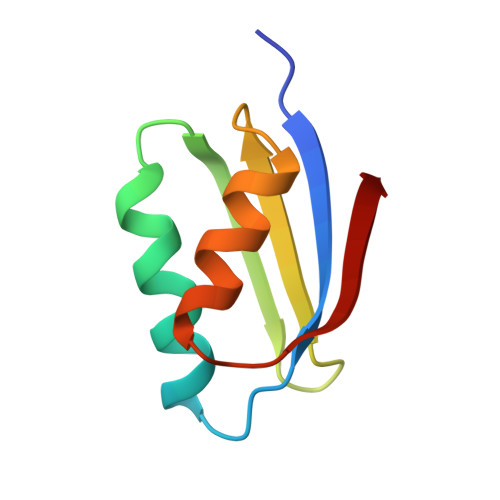
Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Rosenzweig, A.C., Huffman, D.L., Hou, M.Y., Wernimont, A.K., Pufahl, R.A., O'Halloran, T.V.(1999) Structure 7: 605-617
- PubMed: 10404590
- DOI: https://doi.org/10.1016/s0969-2126(99)80082-3
- Primary Citation of Related Structures:
1CC7, 1CC8 - PubMed Abstract:
Metallochaperone proteins function in the trafficking and delivery of essential, yet potentially toxic, metal ions to distinct locations and particular proteins in eukaryotic cells. The Atx1 protein shuttles copper to the transport ATPase Ccc2 in yeast cells. Molecular mechanisms for copper delivery by Atx1 and similar human chaperones have been proposed, but detailed structural characterization is necessary to elucidate how Atx1 binds metal ions and how it might interact with Ccc2 to facilitate metal ion transfer. The 1.02 A resolution X-ray structure of the Hg(II) form of Atx1 (HgAtx1) reveals the overall secondary structure, the location of the metal-binding site, the detailed coordination geometry for Hg(II), and specific amino acid residues that may be important in interactions with Ccc2. Metal ion transfer experiments establish that HgAtx1 is a functional model for the Cu(I) form of Atx1 (CuAtx1). The metal-binding loop is flexible, changing conformation to form a disulfide bond in the oxidized apo form, the structure of which has been solved to 1.20 A resolution. The Atx1 structure represents the first structure of a metallochaperone protein, and is one of the largest unknown structures solved by direct methods. The structural features of the metal-binding site support the proposed Atx1 mechanism in which facile metal ion transfer occurs between metal-binding sites of the diffusible copper-donor and membrane-tethered copper-acceptor proteins. The Atx1 structural motif represents a prototypical metal ion trafficking unit that is likely to be employed in a variety of organisms for different metal ions.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Cell Biology, Northwestern University, Evanston, IL 60208, USA. amyr@nwu.edu