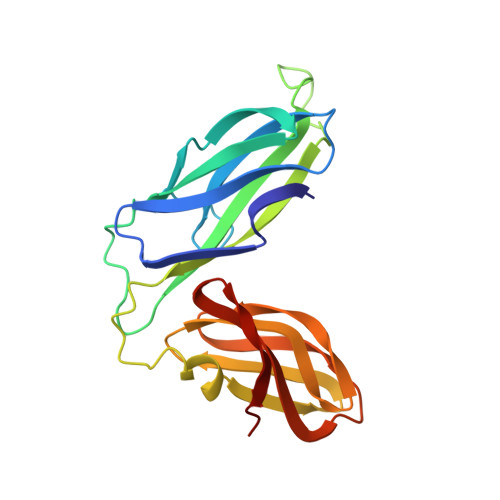
NMR solution structure of the periplasmic chaperone FimC.
Pellecchia, M., Guntert, P., Glockshuber, R., Wuthrich, K.(1998) Nat Struct Biol 5: 885-890
- PubMed: 9783748
- DOI: https://doi.org/10.1038/2325
- Primary Citation of Related Structures:
1BF8 - PubMed Abstract:
The NMR structure of the 205-residue periplasmic chaperone FimC is presented. This protein consists of two globular domains with immunoglobulin-like folds connected by a 15-residue linker peptide. The relative orientation of the two domains is defined by hydrophobic contacts and an interdomain salt bridge. FimC mediates the assembly of type-1 pili, which are filamentous surface organelles of uropathogenic Escherichia coli strains that enable the bacteria to attach to host cell surfaces and persist in macrophages. The availability of the NMR structure of FimC provides a new basis for rational design of drugs against infections by uropathogenic bacteria.
Organizational Affiliation:
Institut für Molekularbiologie und Biophysik, Eidgenössische Technische Hochschule Hönggerberg, Zürich, Switzerland.