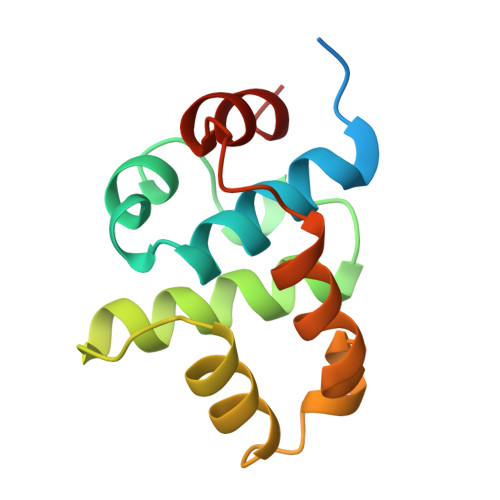
Crystal structure of the N-terminal domain of the DnaB hexameric helicase.
Fass, D., Bogden, C.E., Berger, J.M.(1999) Structure 7: 691-698
- PubMed: 10404598
- DOI: https://doi.org/10.1016/s0969-2126(99)80090-2
- Primary Citation of Related Structures:
1B79 - PubMed Abstract:
The hexameric helicase DnaB unwinds the DNA duplex at the Escherichia coli chromosome replication fork. Although the mechanism by which DnaB both couples ATP hydrolysis to translocation along DNA and denatures the duplex is unknown, a change in the quaternary structure of the protein involving dimerization of the N-terminal domain has been observed and may occur during the enzymatic cycle. This N-terminal domain is required both for interaction with other proteins in the primosome and for DnaB helicase activity. Knowledge of the structure of this domain may contribute to an understanding of its role in DnaB function. We have determined the structure of the N-terminal domain of DnaB crystallographically. The structure is globular, highly helical and lacks a close structural relative in the database of known protein folds. Conserved residues and sites of dominant-negative mutations have structurally significant roles. Each asymmetric unit in the crystal contains two independent and identical copies of a dimer of the DnaB N-terminal domain. The large-scale domain or subunit reorientation that is seen in DnaB by electron microscopy might result from the formation of a true twofold symmetric dimer of N-terminal domains, while maintaining a head-to-tail arrangement of C-terminal domains. The N-terminal domain of DnaB is the first region of a hexameric DNA replicative helicase to be visualized at high resolution. Comparison of this structure to the analogous region of the Rho RNA/DNA helicase indicates that the N-terminal domains of these hexameric helicases are structurally dissimilar.
Organizational Affiliation:
Whitehead Institute, Cambridge, Massachusetts 02142, USA.