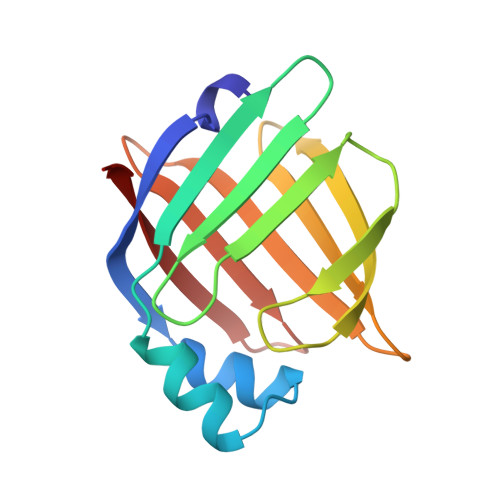
Crystal structure of recombinant murine adipocyte lipid-binding protein.
Xu, Z., Bernlohr, D.A., Banaszak, L.J.(1992) Biochemistry 31: 3484-3492
- PubMed: 1554730
- DOI: https://doi.org/10.1021/bi00128a024
- Primary Citation of Related Structures:
1ALB - PubMed Abstract:
Adipocyte lipid-binding protein (ALBP) is the adipocyte member of an intracellular hydrophobic ligand-binding protein family. ALBP is phosphorylated by the insulin receptor kinase upon insulin stimulation. The crystal structure of recombinant murine ALBP has been determined and refined to 2.5 A. The final R factor for the model is 0.18 with good canonical properties. Crystalline ALBP has a conformation which is essentially identical to that of intestinal fatty acid binding protein and myelin P2 protein. Although the crystal structure is of the apo- form, a cavity resembling that in other family members is present. It contains a number of bound and implied unbound water molecules and shows no large obvious portal to the external milieu. The cavity of ALBP, which by homology is the ligand-binding site, is formed by both polar and hydrophobic residues among which is tyrosine 19. Y19 is phosphorylated by the insulin receptor kinase as described in the accompanying paper [Buelt, M. K., Xu, Z., Banaszak, L. J., & Bernlohr, D. A. (1992) Biochemistry (following paper in this issue)]. By comparing ALBP with the earlier structural results on intestinal fatty acid binding protein, it is now possible to delineate conserved amino acids which help form the binding site in this family.
Organizational Affiliation:
Department of Biochemistry, University of Minnesota Medical School, Minneapolis 55455.