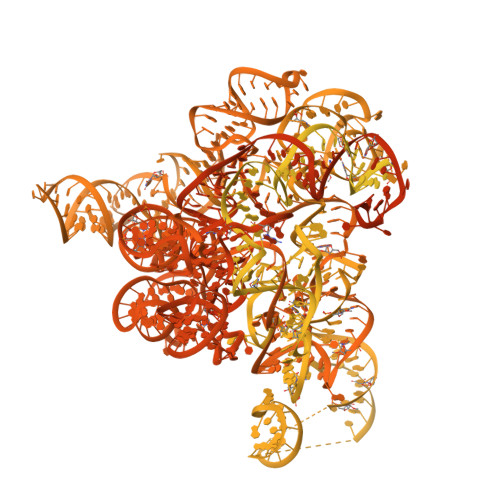
Cryo-EM structure and rRNA modification sites of a plant ribosome.
Cottilli, P., Itoh, Y., Nobe, Y., Petrov, A.S., Lison, P., Taoka, M., Amunts, A.(2022) Plant Commun 3: 100342-100342
- PubMed: 35643637
- DOI: https://doi.org/10.1016/j.xplc.2022.100342
- Primary Citation of Related Structures:
7QIW, 7QIX, 7QIY, 7QIZ - PubMed Abstract:
Protein synthesis in crop plants contributes to the balance of food and fuel on our planet, which influences human metabolic activity and lifespan. Protein synthesis can be regulated with respect to changing environmental cues via the deposition of chemical modifications into rRNA. Here, we present the structure of a plant ribosome from tomato and a quantitative mass spectrometry analysis of its rRNAs. The study reveals fine features of the ribosomal proteins and 71 plant-specific rRNA modifications, and it re-annotates 30 rRNA residues in the available sequence. At the protein level, isoAsp is found in position 137 of uS11, and a zinc finger previously believed to be universal is missing from eL34, suggesting a lower effect of zinc deficiency on protein synthesis in plants. At the rRNA level, the plant ribosome differs markedly from its human counterpart with respect to the spatial distribution of modifications. Thus, it represents an additional layer of gene expression regulation, highlighting the molecular signature of a plant ribosome. The results provide a reference model of a plant ribosome for structural studies and an accurate marker for molecular ecology.
Organizational Affiliation:
Science for Life Laboratory, Department of Biochemistry and Biophysics, Stockholm University, 17165 Solna, Sweden.