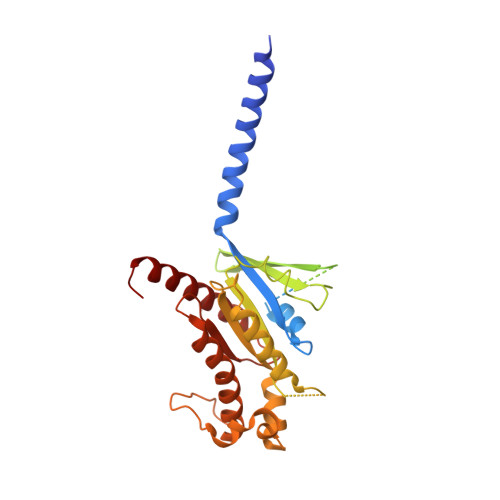
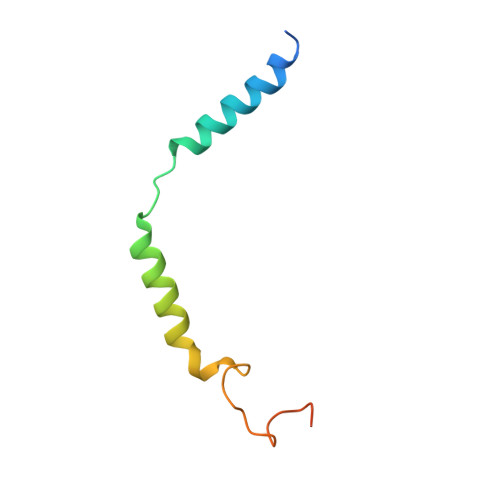
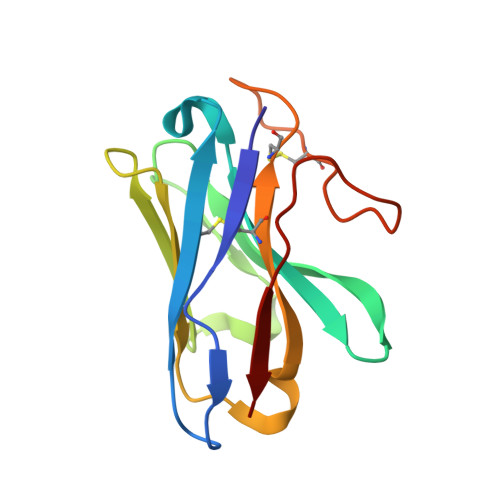
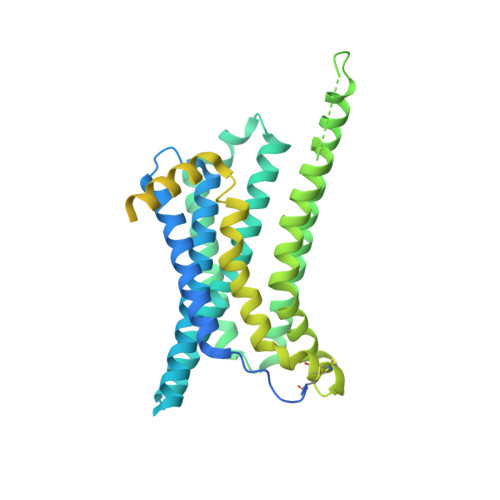
Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Zhang, H., Chen, L.N., Yang, D., Mao, C., Shen, Q., Feng, W., Shen, D.D., Dai, A., Xie, S., Zhou, Y., Qin, J., Sun, J.P., Scharf, D.H., Hou, T., Zhou, T., Wang, M.W., Zhang, Y.(2021) Cell Res 31: 1163-1175
- PubMed: 34433901
- DOI: https://doi.org/10.1038/s41422-021-00552-3
- Primary Citation of Related Structures:
7F53, 7F54, 7F55, 7F58 - PubMed Abstract:
Melanocortin-4 receptor (MC4R) plays a central role in the regulation of energy homeostasis. Its high sequence similarity to other MC receptor family members, low agonist selectivity and the lack of structural information concerning MC4R-specific activation have hampered the development of MC4R-seletive therapeutics to treat obesity. Here, we report four high-resolution structures of full-length MC4R in complex with the heterotrimeric G s protein stimulated by the endogenous peptide ligand α-MSH, FDA-approved drugs afamelanotide (Scenesse™) and bremelanotide (Vyleesi™), and a selective small-molecule ligand THIQ, respectively. Together with pharmacological studies, our results reveal the conserved binding mode of peptidic agonists, the distinctive molecular details of small-molecule agonist recognition underlying receptor subtype selectivity, and a distinct activation mechanism for MC4R, thereby offering new insights into G protein coupling. Our work may facilitate the discovery of selective therapeutic agents targeting MC4R.
Organizational Affiliation:
Department of Biophysics and Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China.