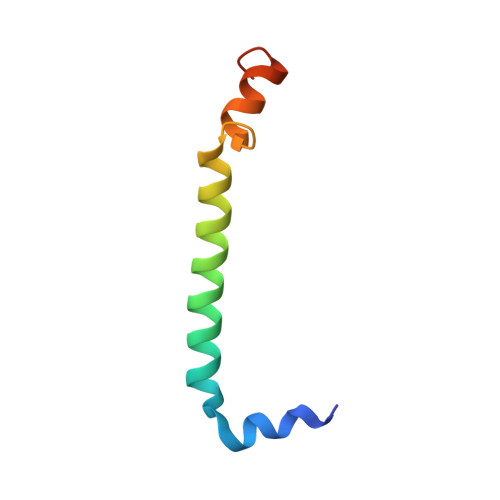
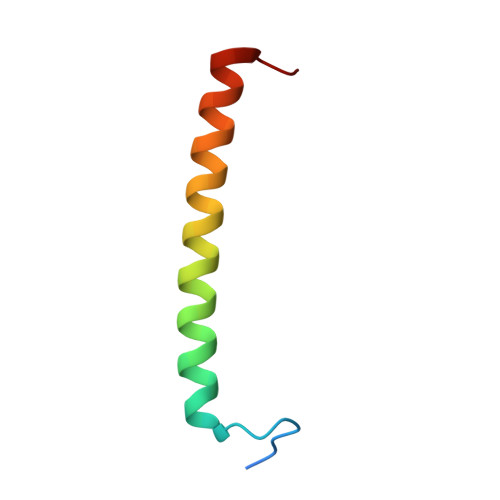
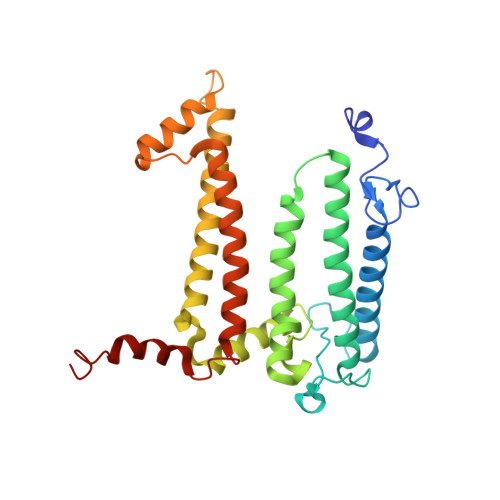
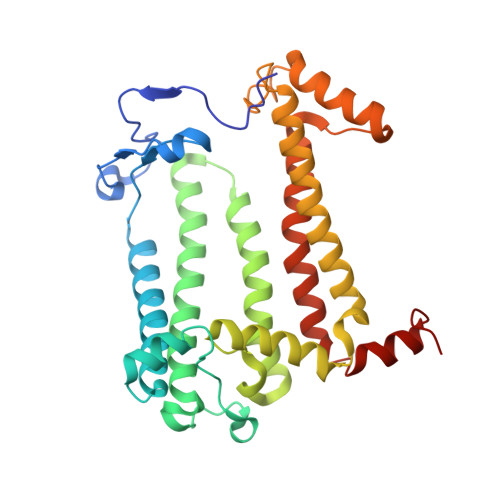
Cryo-EM structure of the photosynthetic RC-LH1-PufX supercomplex at 2.8-angstrom resolution.
Bracun, L., Yamagata, A., Christianson, B.M., Terada, T., Canniffe, D.P., Shirouzu, M., Liu, L.N.(2021) Sci Adv 7
- PubMed: 34134992
- DOI: https://doi.org/10.1126/sciadv.abf8864
- Primary Citation of Related Structures:
7DDQ - PubMed Abstract:
The reaction center (RC)-light-harvesting complex 1 (LH1) supercomplex plays a pivotal role in bacterial photosynthesis. Many RC-LH1 complexes integrate an additional protein PufX that is key for bacterial growth and photosynthetic competence. Here, we present a cryo-electron microscopy structure of the RC-LH1-PufX supercomplex from Rhodobacter veldkampii at 2.8-Å resolution. The RC-LH1-PufX monomer contains an LH ring of 15 αβ-polypeptides with a 30-Å gap formed by PufX. PufX acts as a molecular "cross brace" to reinforce the RC-LH1 structure. The unusual PufX-mediated large opening in the LH1 ring and defined arrangement of proteins and cofactors provide the molecular basis for the assembly of a robust RC-LH1-PufX supercomplex and efficient quinone transport and electron transfer. These architectural features represent the natural strategies for anoxygenic photosynthesis and environmental adaptation.
Organizational Affiliation:
Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.