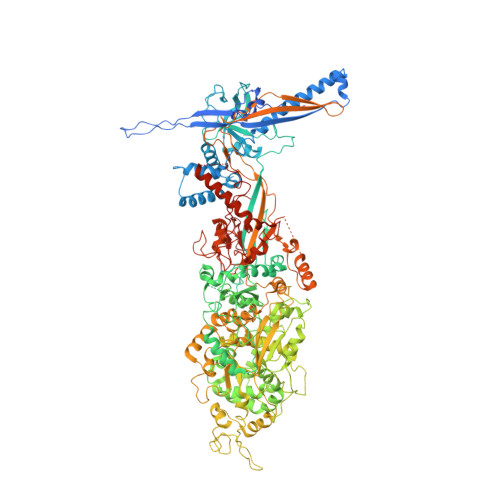
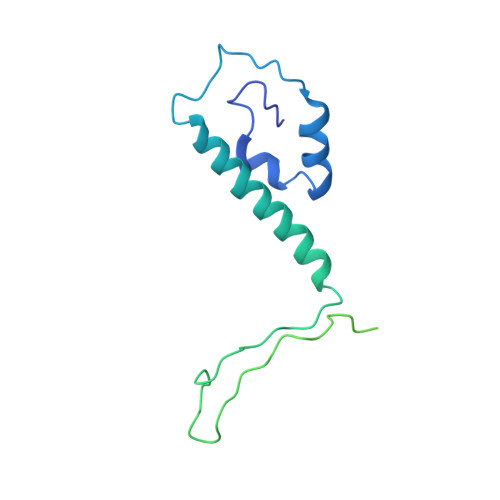
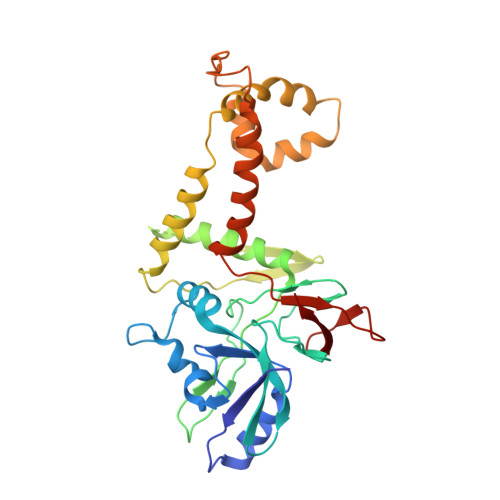
Cryo-EM structure of the varicella-zoster virus A-capsid.
Sun, J., Liu, C., Peng, R., Zhang, F.K., Tong, Z., Liu, S., Shi, Y., Zhao, Z., Zeng, W.B., Gao, G.F., Shen, H.J., Yang, X., Luo, M., Qi, J., Wang, P.(2020) Nat Commun 11: 4795-4795
- PubMed: 32963252
- DOI: https://doi.org/10.1038/s41467-020-18537-y
- Primary Citation of Related Structures:
7BW6 - PubMed Abstract:
Varicella-zoster virus (VZV), a member of the Alphaherpesvirinae subfamily, causes severe diseases in humans of all ages. The viral capsids play critical roles in herpesvirus infection, making them potential antiviral targets. Here, we present the 3.7-Å-resolution structure of the VZV A-capsid and define the molecular determinants underpinning the assembly of this complicated viral machinery. Overall, the VZV capsid has a similar architecture to that of other known herpesviruses. The major capsid protein (MCP) assembles into pentons and hexons, forming extensive intra- and inter-capsomer interaction networks that are further secured by the small capsid protein (SCP) and the heterotriplex. The structure reveals a pocket beneath the floor of MCP that could potentially be targeted by antiviral inhibitors. In addition, we identified two alphaherpesvirus-specific structural features in SCP and Tri1 proteins. These observations highlight the divergence of different herpesviruses and provide an important basis for developing antiviral drugs.
Organizational Affiliation:
Shanxi Academy of Advanced Research and Innovation, Taiyuan, 030032, China.