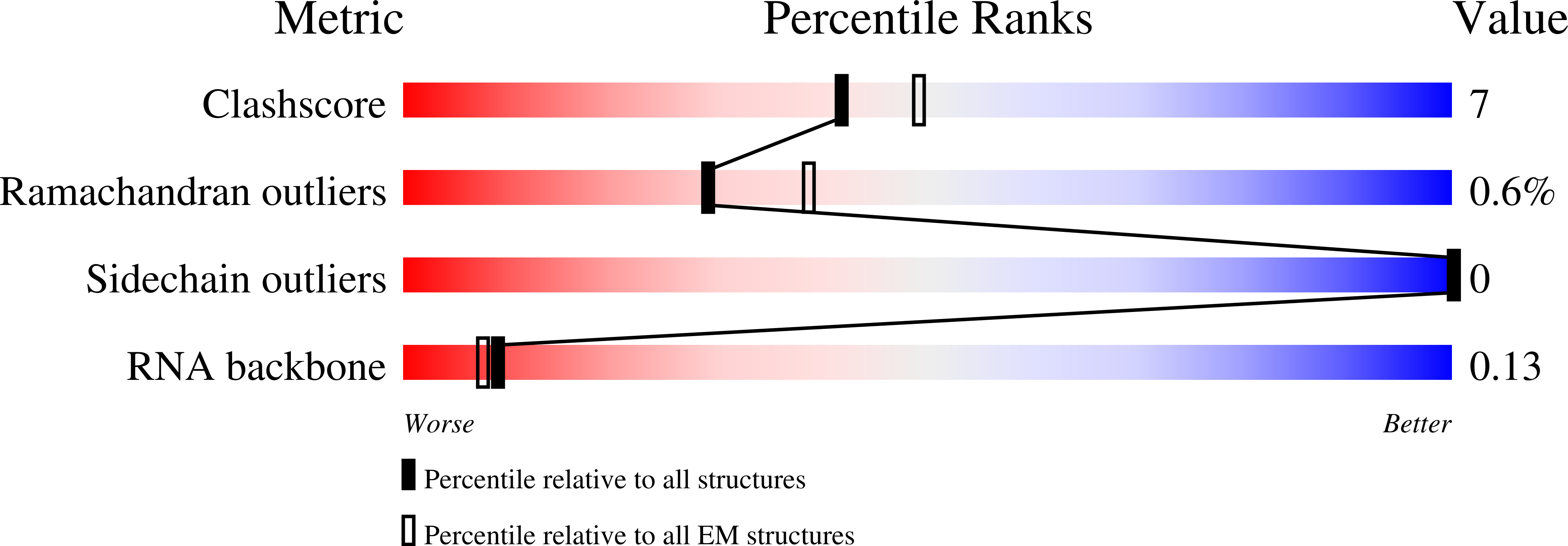
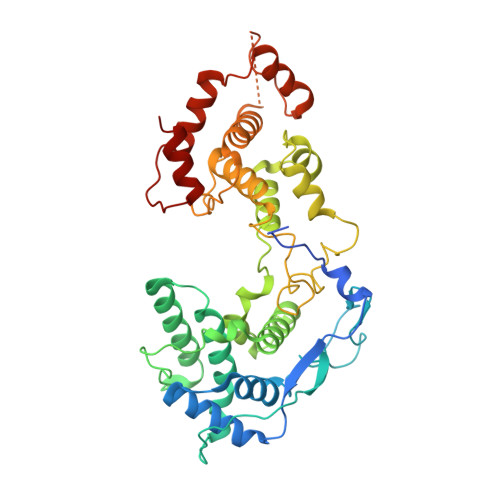
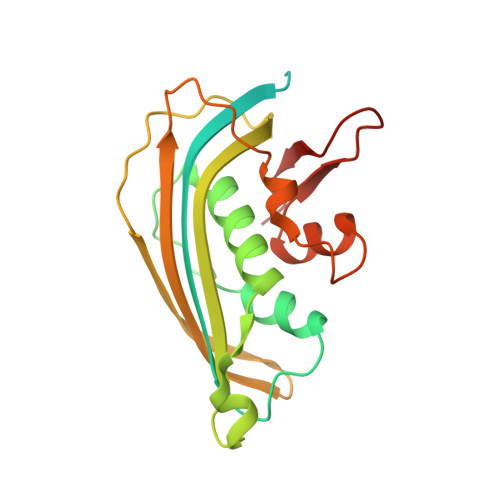
Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle.
Jenni, S., Horwitz, J.A., Bloyet, L.M., Whelan, S.P.J., Harrison, S.C.(2022) Nat Commun 13: 4802-4802
- PubMed: 35970826
- DOI: https://doi.org/10.1038/s41467-022-32223-1
- Primary Citation of Related Structures:
7UMK, 7UML - PubMed Abstract:
Vesicular stomatitis virus (VSV) is a negative-strand RNA virus with a non-segmented genome, closely related to rabies virus. Both have characteristic bullet-like shapes. We report the structure of intact, infectious VSV particles determined by cryogenic electron microscopy. By compensating for polymorphism among viral particles with computational classification, we obtained a reconstruction of the shaft ("trunk") at 3.5 Å resolution, with lower resolution for the rounded tip. The ribonucleoprotein (RNP), genomic RNA complexed with nucleoprotein (N), curls into a dome-like structure with about eight gradually expanding turns before transitioning into the regular helical trunk. Two layers of matrix (M) protein link the RNP with the membrane. Radial inter-layer subunit contacts are fixed within single RNA-N-M1-M2 modules, but flexible lateral and axial interactions allow assembly of polymorphic virions. Together with published structures of recombinant N in various states, our results suggest a mechanism for membrane-coupled self-assembly of VSV and its relatives.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, 02115, USA. jenni@crystal.harvard.edu.