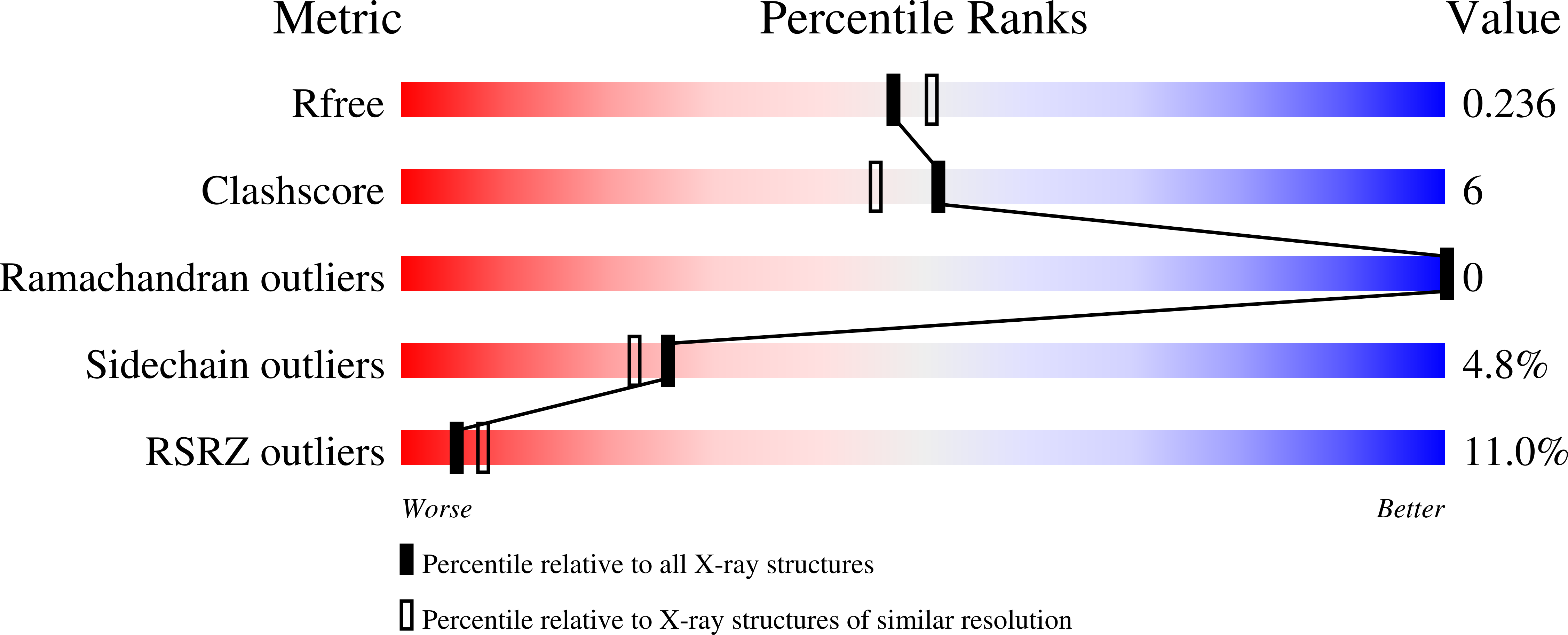
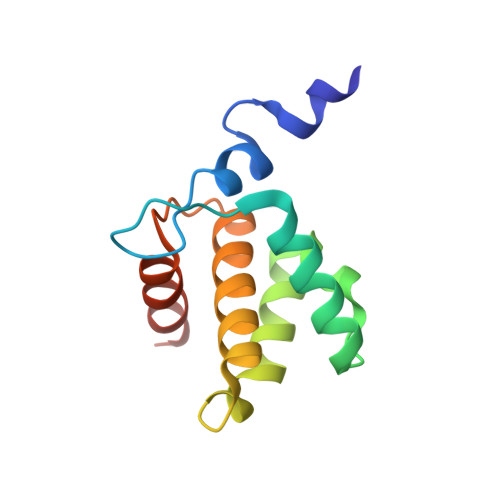
Atomic view of the HIV-1 matrix lattice; implications on virus assembly and envelope incorporation.
Samal, A.B., Green, T.J., Saad, J.S.(2022) Proc Natl Acad Sci U S A 119: e2200794119-e2200794119
- PubMed: 35658080
- DOI: https://doi.org/10.1073/pnas.2200794119
- Primary Citation of Related Structures:
7TBP - PubMed Abstract:
During the late phase of HIV type 1 (HIV-1) infection cycle, the virally encoded Gag polyproteins are targeted to the inner leaflet of the plasma membrane (PM) for assembly, formation of immature particles, and virus release. Gag binding to the PM is mediated by interactions of the N-terminally myristoylated matrix (myrMA) domain with phosphatidylinositol 4,5-bisphosphate. Formation of a myrMA lattice on the PM is an obligatory step for the assembly of immature HIV-1 particles and envelope (Env) incorporation. Atomic details of the myrMA lattice and how it mediates Env incorporation are lacking. Herein, we present the X-ray structure of myrMA at 2.15 Å. The myrMA lattice is arranged as a hexamer of trimers with a central hole, thought to accommodate the C-terminal tail of Env to promote incorporation into virions. The trimer–trimer interactions in the lattice are mediated by the N-terminal loop of one myrMA molecule and α-helices I–II, as well as the 310 helix of a myrMA molecule from an adjacent trimer. We provide evidence that substitution of MA residues Leu13 and Leu31, previously shown to have adverse effects on Env incorporation, induced a conformational change in myrMA, which may destabilize the trimer–trimer interactions within the lattice. We also show that PI(4,5)P2 is capable of binding to alternating sites on MA, consistent with an MA–membrane binding mechanism during assembly of the immature particle and upon maturation. Altogether, these findings advance our understanding of a key mechanism in HIV-1 particle assembly.
Organizational Affiliation:
Department of Microbiology, University of Alabama at Birmingham, Birmingham, AL 35294.