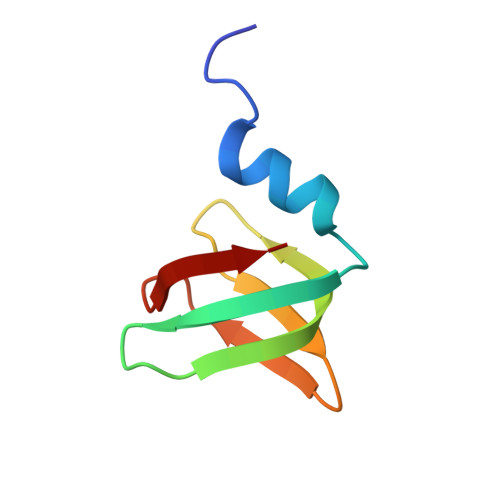
Structure and RNA-Binding Properties of Lsm Protein from Halobacterium salinarum.
Fando, M.S., Mikhaylina, A.O., Lekontseva, N.V., Tishchenko, S.V., Nikulin, A.D.(2021) Biochemistry (Mosc) 86: 833-842
- PubMed: 34284708
- DOI: https://doi.org/10.1134/S000629792107004X
- Primary Citation of Related Structures:
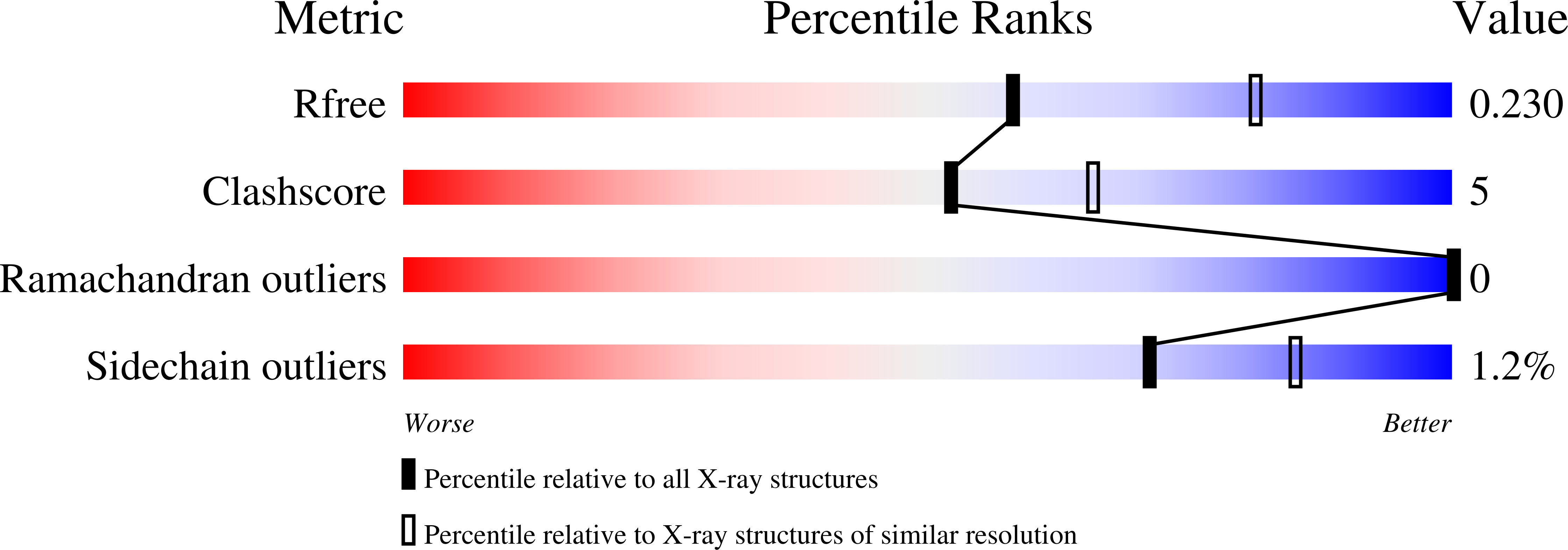
6TFL - PubMed Abstract:
The structure and the RNA-binding properties of the Lsm protein from Halobacterium salinarum have been determined. A distinctive feature of this protein is the presence of a short L4 loop connecting the β3 and β4 strands. Since bacterial Lsm proteins (also called Hfq proteins) have a short L4 loop and form hexamers, whereas archaeal Lsm proteins (SmAP) have a long L4 loop and form heptamers, it has been suggested that the length of the L4 loop may affect the quaternary structure of Lsm proteins. Moreover, the L4 loop covers the region of SmAP corresponding to one of the RNA-binding sites in Hfq, and thus can affect the RNA-binding properties of the protein. Our results show that the SmAP from H. salinarum forms heptamers and possesses the same RNA-binding properties as homologous proteins with the long L4 loop. Therefore, the length of the L4 does not govern the number of monomers in the protein particles and does not affect the RNA-binding properties of Lsm proteins.
Organizational Affiliation:
Institute of Protein Research Russian Academy of Sciences, Pushchino, Moscow Region, 142290, Russia.