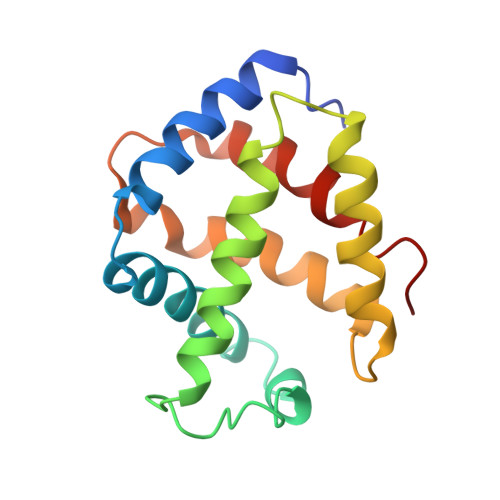
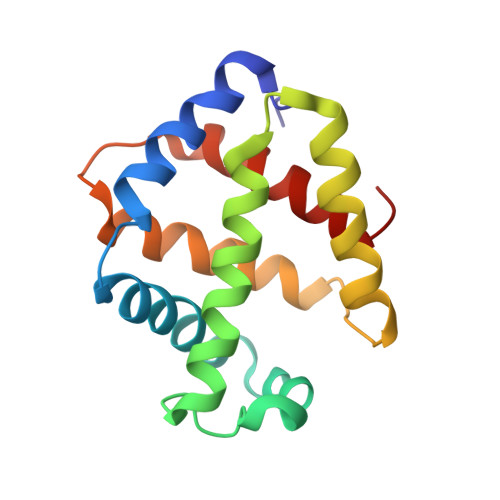
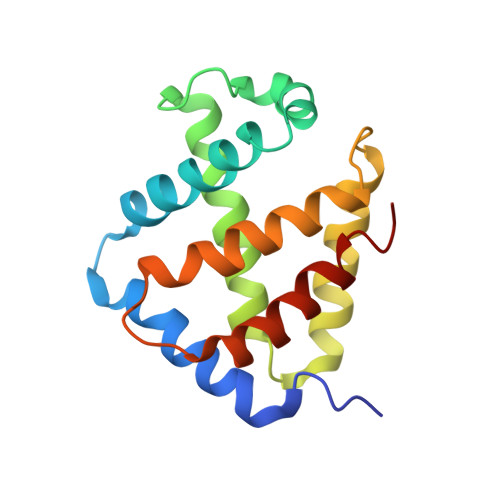
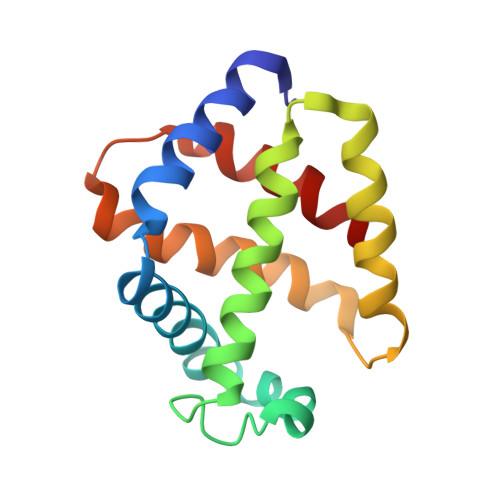
Single-particle cryo-EM using alignment by classification (ABC): the structure of Lumbricus terrestris haemoglobin.
Afanasyev, P., Seer-Linnemayr, C., Ravelli, R.B.G., Matadeen, R., De Carlo, S., Alewijnse, B., Portugal, R.V., Pannu, N.S., Schatz, M., van Heel, M.(2017) IUCrJ 4: 678-694
- PubMed: 28989723
- DOI: https://doi.org/10.1107/S2052252517010922
- Primary Citation of Related Structures:
5M3L - PubMed Abstract:
Single-particle cryogenic electron microscopy (cryo-EM) can now yield near-atomic resolution structures of biological complexes. However, the reference-based alignment algorithms commonly used in cryo-EM suffer from reference bias, limiting their applicability (also known as the 'Einstein from random noise' problem). Low-dose cryo-EM therefore requires robust and objective approaches to reveal the structural information contained in the extremely noisy data, especially when dealing with small structures. A reference-free pipeline is presented for obtaining near-atomic resolution three-dimensional reconstructions from heterogeneous ('four-dimensional') cryo-EM data sets. The methodologies integrated in this pipeline include a posteriori camera correction, movie-based full-data-set contrast transfer function determination, movie-alignment algorithms, (Fourier-space) multivariate statistical data compression and unsupervised classification, 'random-startup' three-dimensional reconstructions, four-dimensional structural refinements and Fourier shell correlation criteria for evaluating anisotropic resolution. The procedures exclusively use information emerging from the data set itself, without external 'starting models'. Euler-angle assignments are performed by angular reconstitution rather than by the inherently slower projection-matching approaches. The comprehensive 'ABC-4D' pipeline is based on the two-dimensional reference-free 'alignment by classification' (ABC) approach, where similar images in similar orientations are grouped by unsupervised classification. Some fundamental differences between X-ray crystallography versus single-particle cryo-EM data collection and data processing are discussed. The structure of the giant haemoglobin from Lumbricus terrestris at a global resolution of ∼3.8 Å is presented as an example of the use of the ABC-4D procedure.
Organizational Affiliation:
Institute of Biology Leiden, Leiden University, 2333 CC Leiden, The Netherlands.