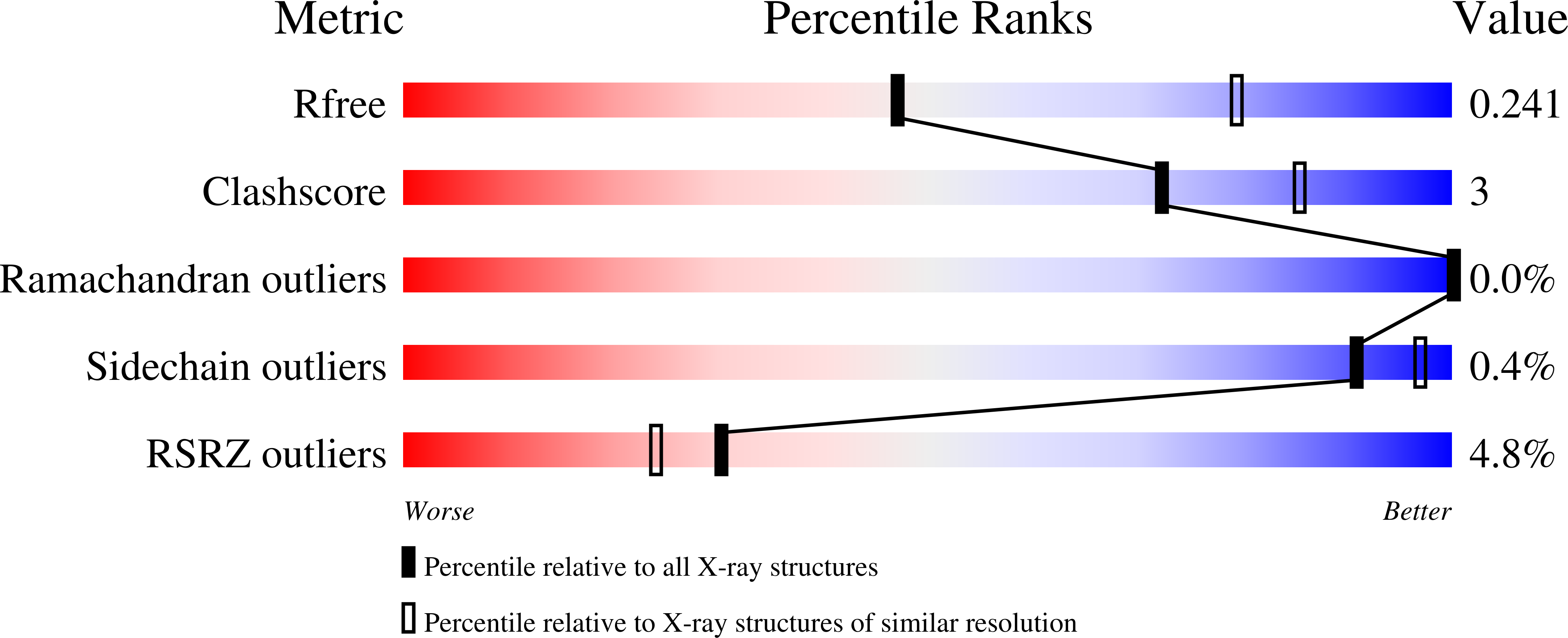
Structural Studies of AAV2 Rep68 Reveal a Partially Structured Linker and Compact Domain Conformation.
Musayev, F.N., Zarate-Perez, F., Bardelli, M., Bishop, C., Saniev, E.F., Linden, R.M., Henckaerts, E., Escalante, C.R.(2015) Biochemistry 54: 5907-5919
- PubMed: 26314310
- DOI: https://doi.org/10.1021/acs.biochem.5b00610
- Primary Citation of Related Structures:
4ZO0, 5DCX - PubMed Abstract:
Adeno-associated virus (AAV) nonstructural proteins Rep78 and Rep68 carry out all DNA transactions that regulate the AAV life cycle. They share two multifunctional domains: an N-terminal origin binding/nicking domain (OBD) from the HUH superfamily and a SF3 helicase domain. A short linker of ∼20 amino acids that is critical for oligomerization and function connects the two domains. Although X-ray structures of the AAV5 OBD and AAV2 helicase domains have been determined, information about the full-length protein and linker conformation is not known. This article presents the solution structure of AAV2 Rep68 using small-angle X-ray scattering (SAXS). We first determined the X-ray structures of the minimal AAV2 Rep68 OBD and of the OBD with the linker region. These X-ray structures reveal novel features that include a long C-terminal α-helix that protrudes from the core of the protein at a 45° angle and a partially structured linker. SAXS studies corroborate that the linker is not extended, and we show that a proline residue in the linker is critical for Rep68 oligomerization and function. SAXS-based rigid-body modeling of Rep68 confirms these observations, showing a compact arrangement of the two domains in which they acquire a conformation that positions key residues in all domains on one face of the protein, poised to interact with DNA.
Organizational Affiliation:
Department of Infectious Diseases, King's College London , London SE1 9RT, United Kingdom.