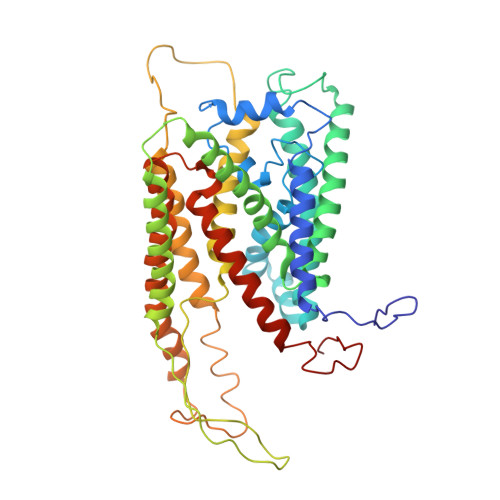
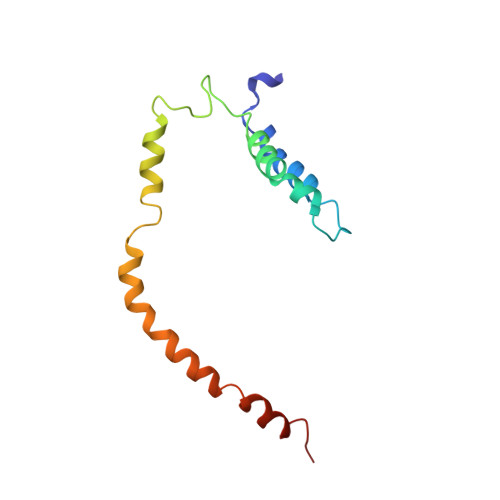
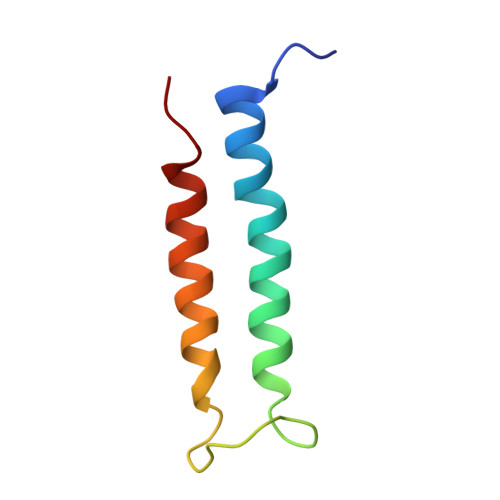
Visualization of a Polytopic Membrane Protein During Secy-Mediated Membrane Insertion.
Bischoff, L., Wickles, S., Berninghausen, O., Van Der Sluis, E.O., Beckmann, R.(2014) Nat Commun 5: 4103
- PubMed: 24912953
- DOI: https://doi.org/10.1038/ncomms5103
- Primary Citation of Related Structures:
5ABB - PubMed Abstract:
The biogenesis of polytopic membrane proteins occurs co-translationally on ribosomes that are tightly bound to a membrane-embedded protein-conducting channel: the Sec-complex. The path that is followed by nascent proteins inside the ribosome and the Sec-complex is relatively well established; however, it is not clear what the fate of the N-terminal transmembrane domains (TMDs) of polytopic membrane proteins is when the C-terminal TMDs domains are not yet synthesized. Here, we present the sub-nanometer cryo-electron microscopy structure of an in vivo generated ribosome-SecY complex that carries a membrane insertion intermediate of proteorhodopsin (PR). The structure reveals a pre-opened Sec-complex and the first two TMDs of PR already outside the SecY complex directly in front of its proposed lateral gate. Thus, our structure is in agreement with positioning of N-terminal TMDs at the periphery of SecY, and in addition, it provides clues for the molecular mechanism underlying membrane protein topogenesis.
Organizational Affiliation:
Department of Biochemistry, Gene Center and Center for integrated Protein Science Munich, Feodor-Lynen-Strasse 25, University of Munich, 81377 Munich, Germany.