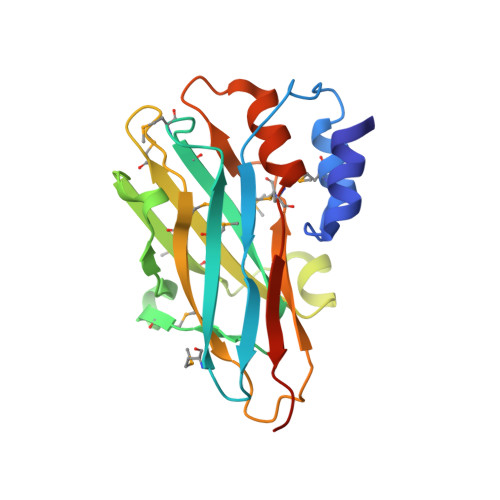
Structural characterization of a novel autonomous cohesin from Ruminococcus flavefaciens.
Voronov-Goldman, M., Levy-Assaraf, M., Yaniv, O., Wisserman, G., Jindou, S., Borovok, I., Bayer, E.A., Lamed, R., Shimon, L.J., Frolow, F.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 450-456
- PubMed: 24699736
- DOI: https://doi.org/10.1107/S2053230X14004051
- Primary Citation of Related Structures:
4N2O - PubMed Abstract:
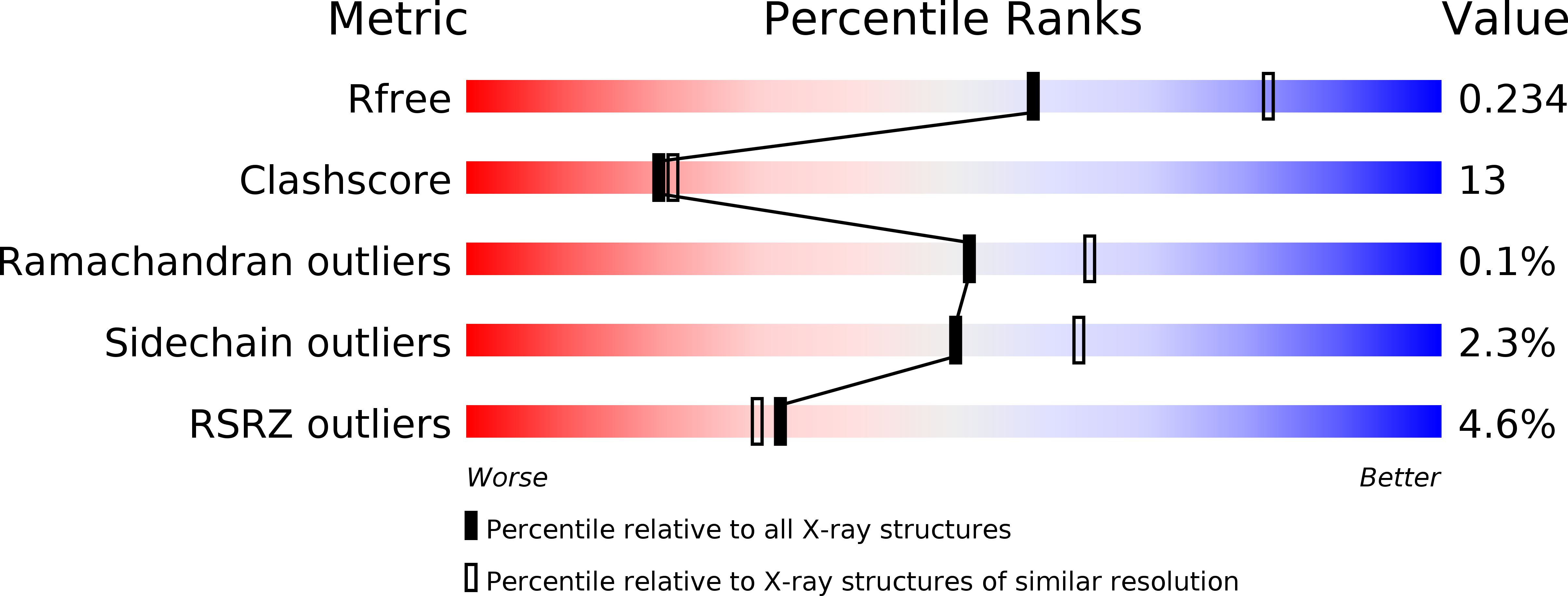
Ruminococcus flavefaciens is a cellulolytic bacterium found in the rumen of herbivores and produces one of the most elaborate and variable cellulosome systems. The structure of an R. flavefaciens protein (RfCohG, ZP_06142108), representing a freestanding (non-cellulosomal) type III cohesin module, has been determined. A selenomethionine derivative with a C-terminal histidine tag was crystallized and diffraction data were measured to 2.44 Å resolution. Its structure was determined by single-wavelength anomalous dispersion, revealing eight molecules in the asymmetric unit. RfCohG exhibits the most complex among all known cohesin structures, possessing four α-helical elements and a topographical protuberance on the putative dockerin-binding surface.
Organizational Affiliation:
Department of Molecular Microbiology and Biotechnology, Tel Aviv University, Tel Aviv 69978, Israel.