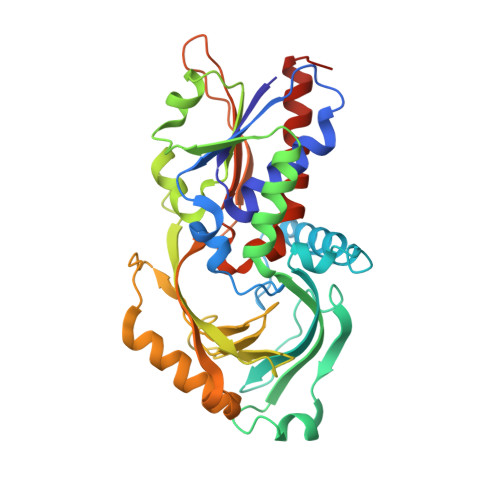
Three-dimensional structure of porcine kidney D-amino acid oxidase at 3.0 A resolution.
Mizutani, H., Miyahara, I., Hirotsu, K., Nishina, Y., Shiga, K., Setoyama, C., Miura, R.(1996) J Biochem 120: 14-17
- PubMed: 8864836
- DOI: https://doi.org/10.1093/oxfordjournals.jbchem.a021376
- Primary Citation of Related Structures:
1VE9 - PubMed Abstract:
The X-ray crystallographic structure of porcine kidney D-amino acid oxidase, which had been expressed in Escherichia coli transformed with a vector containing DAO cDNA, was determined by the isomorphous replacement method for the complex form with benzoate. The known amino acid sequence, FAD and benzoate were fitted to an electron density map of 3.0 A resolution with an R-factor of 21.0%. The overall dimeric structure exhibits an elongated ellipsoidal framework. The prosthetic group, FAD, was found to be in an extended conformation, the isoalloxazine ring being buried in the protein core. The ADP moiety of FAD was located in the typical beta alpha beta dinucleotide binding motif, with the alpha-helix dipole stabilizing the pyrophosphate negative charge. The substrate analog, benzoate, is located on the re-face of the isoalloxazine ring, while the si-face is blocked by hydrophobic residues. The carboxylate group of benzoate is ion-paired with the Arg283 side chain and is within interacting distance with the hydroxy moiety of Tyr228. The phenol ring of Tyr224 is located just above the benzene ring of benzoate, implying the importance of this residue for catalysis. There is no positive charge or alpha-helix dipole near N(1) of flavin. Hydrogen bonds were observed at C(2) = O, N(3)-H, C(4) = O, and N(5) of the flavin ring.
Organizational Affiliation:
Department of Chemistry, Faculty of Science, Osaka City University.