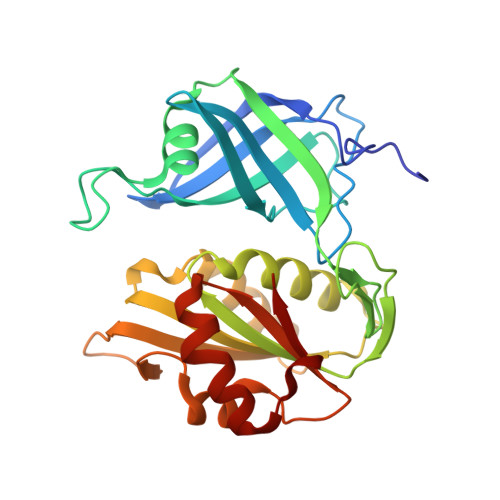
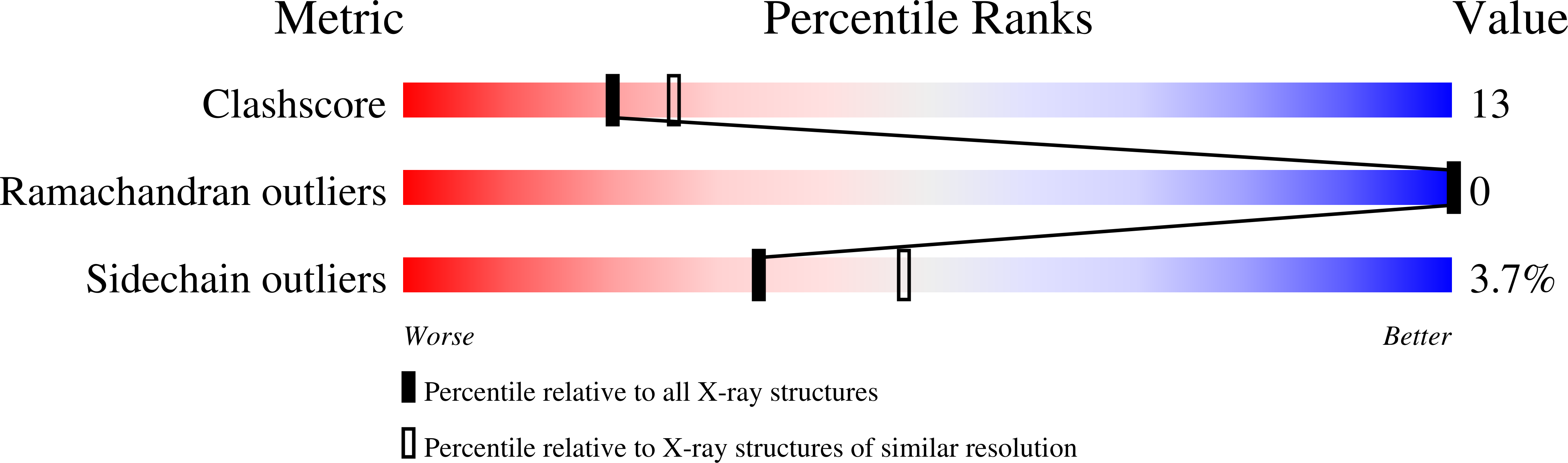The structure and biochemistry of NADH-dependent cytochrome b5 reductase are now consistent.
Bewley, M.C., Marohnic, C.C., Barber, M.J.(2001) Biochemistry 40: 13574-13582
- PubMed: 11695905
- DOI: https://doi.org/10.1021/bi0106336
- Primary Citation of Related Structures:
1I7P, 1IB0 - PubMed Abstract:
Cytochrome b5 reductase (cb5r) (EC 1.6.6.2) catalyzes the reduction of two molecules of cytochrome b5 using NADH as the physiological electron donor. The structure of pig cb5r at 2.4 A resolution was previously reported in the literature, but it was inconsistent with the biochemistry; for example, K83 and C245 were both implicated in the mechanism, but were not located at the active site. To address this problem, we have determined the structures of cb5r from rat at 2.0 A resolution and in a complex with NAD+ at 2.3 A resolution. We found significant differences throughout the rat structure compared to that of pig, including the locations of the lysine and cysteine residues mentioned above. To test the structural models, we made single amino acid substitutions of this lysine and showed that all substitutions produced correctly folded proteins and exhibited normal flavin behavior. However, the apparent kcat(NADH) decreased, and the apparent K(m) for NADH increased; the K(m)'s for cytochrome b5 were unchanged relative to that of the wild type. The largest effect was for the glutamate-substituted protein, which was further characterized using a charge transfer assay and found to be less efficient at NADH utilization than the wild type. These results are consistent with a role for this lysine in stabilizing the NADH-bound form of cb5r. We have concluded that the pig structure was mistraced in several regions and have reinterpreted mutants in these regions that give rise to the hereditary disease methemoglobinemia.
Organizational Affiliation:
Biology Department, Brookhaven National Laboratory, Upton, New York 11973, USA. bewley@bnl.gov