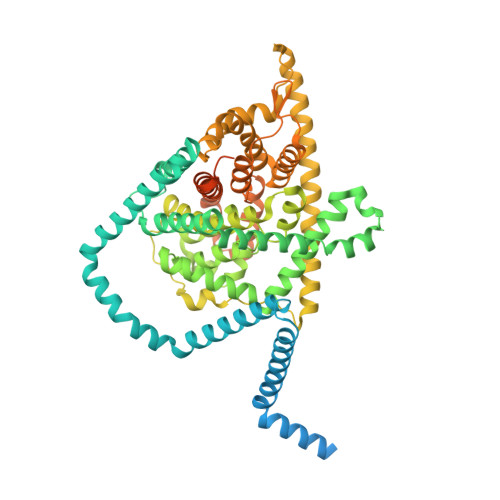
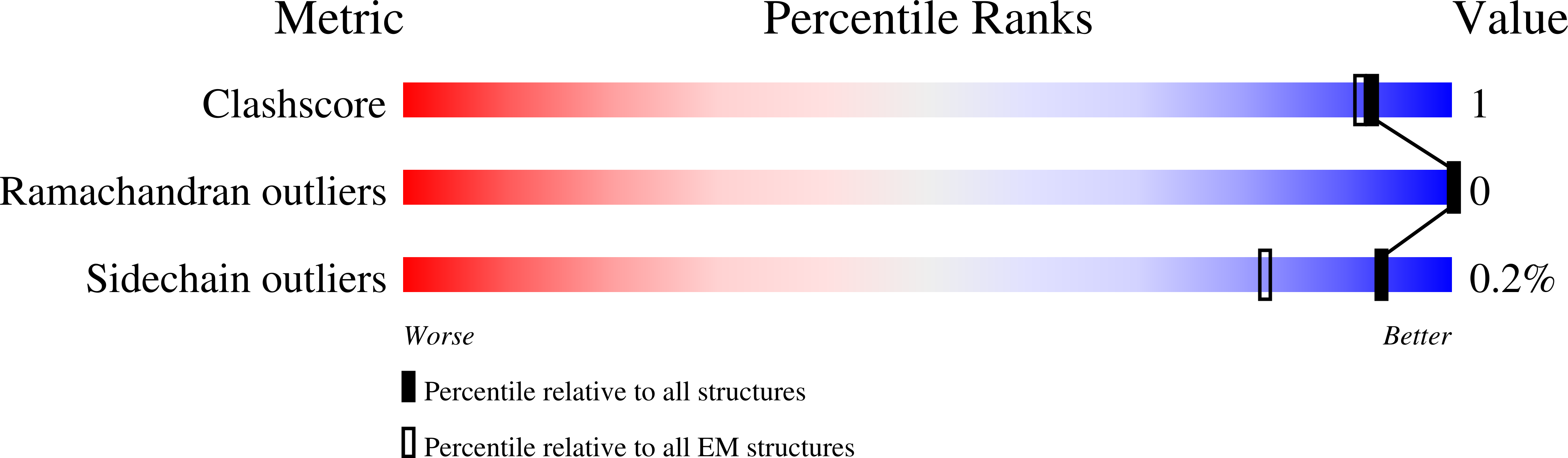Antiviral drug recognition and elevator-type transport motions of CNT3.
Wright, N.J., Zhang, F., Suo, Y., Kong, L., Yin, Y., Fedor, J.G., Sharma, K., Borgnia, M.J., Im, W., Lee, S.Y.(2024) Nat Chem Biol
- PubMed: 38418906
- DOI: https://doi.org/10.1038/s41589-024-01559-8
- Primary Citation of Related Structures:
8TZ1, 8TZ2, 8TZ3, 8TZ4, 8TZ5, 8TZ6, 8TZ7, 8TZ8, 8TZ9, 8TZA, 8TZD - PubMed Abstract:
Nucleoside analogs have broad clinical utility as antiviral drugs. Key to their systemic distribution and cellular entry are human nucleoside transporters. Here, we establish that the human concentrative nucleoside transporter 3 (CNT3) interacts with antiviral drugs used in the treatment of coronavirus infections. We report high-resolution single-particle cryo-electron microscopy structures of bovine CNT3 complexed with antiviral nucleosides N 4 -hydroxycytidine, PSI-6206, GS-441524 and ribavirin, all in inward-facing states. Notably, we found that the orally bioavailable antiviral molnupiravir arrests CNT3 in four distinct conformations, allowing us to capture cryo-electron microscopy structures of drug-loaded outward-facing and drug-loaded intermediate states. Our studies uncover the conformational trajectory of CNT3 during membrane transport of a nucleoside analog antiviral drug, yield new insights into the role of interactions between the transport and the scaffold domains in elevator-like domain movements during drug translocation, and provide insights into the design of nucleoside analog antiviral prodrugs with improved oral bioavailability.
Organizational Affiliation:
Department of Biochemistry, Duke University School of Medicine, Durham, NC, USA.