Crystal Structure of Peptidylprolyl Isomerase from Naegleria fowleri with bound FK506
Dranow, D.M., Bertolin, B.A., Fox III, D., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Currently 6MKE does not have a validation slider image.
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptidylprolyl isomerase | 127 | Naegleria fowleri | Mutation(s): 0 Gene Names: NF0084240 EC: 5.2.1.8 | 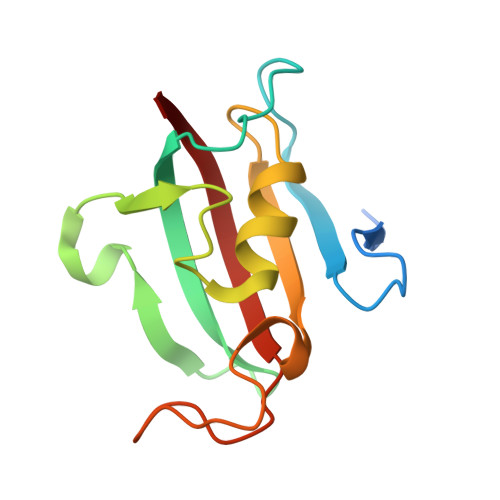 | |
UniProt | |||||
Find proteins for A0A2H4A315 (Naegleria fowleri) Explore A0A2H4A315 Go to UniProtKB: A0A2H4A315 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2H4A315 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FK5 Query on FK5 | E [auth A], F [auth B], G [auth C], H [auth D] | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN C44 H69 N O12 QJJXYPPXXYFBGM-LFZNUXCKSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.05 | α = 90 |
| b = 60.16 | β = 97.12 |
| c = 73.34 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
Currently 6MKE does not have a validation slider image.