Contribution of Physical Interactions to Signaling Specificity between a Diguanylate Cyclase and Its Effector.
Dahlstrom, K.M., Giglio, K.M., Collins, A.J., Sondermann, H., O'Toole, G.A.(2015) mBio 6: e01978-e01915
- PubMed: 26670387
- DOI: https://doi.org/10.1128/mBio.01978-15
- Primary Citation of Related Structures:
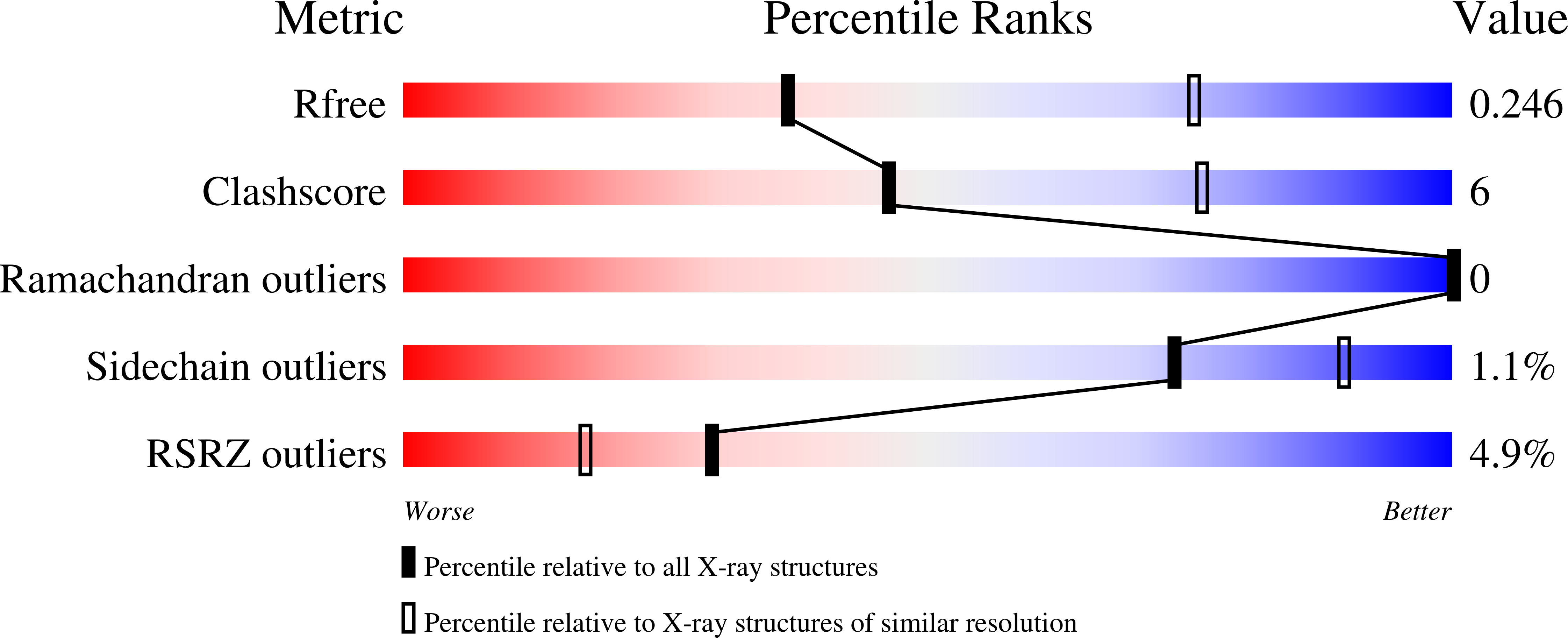
5EUH - PubMed Abstract:
Cyclic diguanylate (c-di-GMP) is a bacterial second messenger that controls multiple cellular processes. c-di-GMP networks have up to dozens of diguanylate cyclases (DGCs) that synthesize c-di-GMP along with many c-di-GMP-responsive target proteins that can bind and respond to this signal. For such networks to have order, a mechanism(s) likely exists that allow DGCs to specifically signal their targets, and it has been suggested that physical interactions might provide such specificity. Our results show a DGC from Pseudomonas fluorescens physically interacting with its target protein at a conserved interface, and this interface can be predictive of DGC-target protein interactions. Furthermore, we demonstrate that physical interaction is necessary for the DGC to maximally signal its target. If such "local signaling" is a theme for even a fraction of the DGCs used by bacteria, it becomes possible to posit a model whereby physical interaction allows a DGC to directly signal its target protein, which in turn may help curtail undesired cross talk with other members of the network.
Organizational Affiliation:
Department of Microbiology and Immunology, Geisel School of Medicine at Dartmouth, Hanover, New Hampshire.