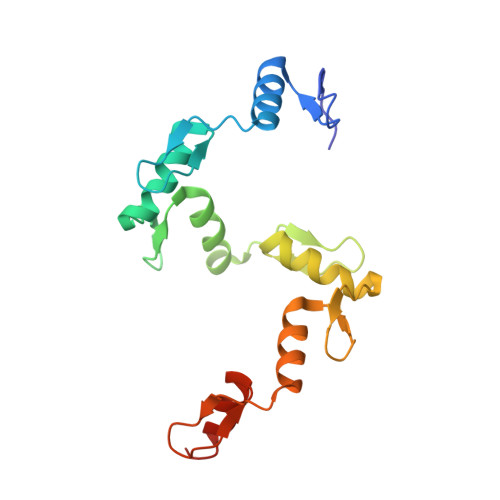
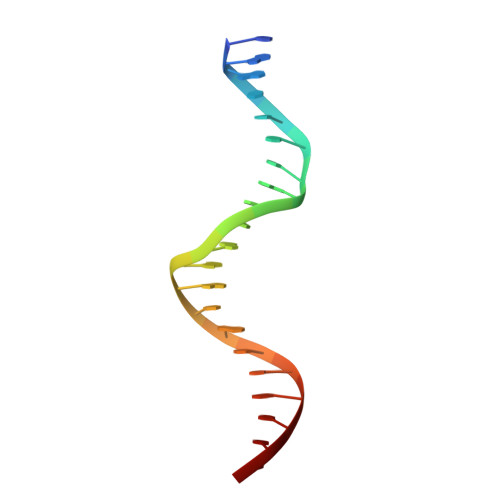
Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence.
Patel, A., Zhang, X., Blumenthal, R.M., Cheng, X.(2017) J Biol Chem 292: 15994-16002
- PubMed: 28801461
- DOI: https://doi.org/10.1074/jbc.M117.805754
- Primary Citation of Related Structures:
5V3G - PubMed Abstract:
PRDM9 is the only mammalian gene that has been associated with speciation. The PR/SET domain 9 (PRDM9) protein is a major determinant of meiotic recombination hot spots and acts through sequence-specific DNA binding via its C2H2 zinc finger (ZF) tandem array, which is highly polymorphic within and between species. The most common human variant, PRDM9 allele A (PRDM9a), contains 13 fingers (ZF1-13). Allele C (PRDM9c) is the second-most common among African populations and differs from PRDM9a by an arginine-to-serine change (R764S) in ZF9 and by replacement of ZF11 with two other fingers, yielding 14 fingers in PRDM9c. Here we co-crystallized the six-finger fragment ZF8-13 of PRDM9c, in complex with an oligonucleotide representing a known PRDM9c-specific hot spot sequence, and compared the structure with that of a characterized PRDM9a-specific complex. There are three major differences. First, Ser 764 in ZF9 allows PRDM9c to accommodate a variable base, whereas PRDM9a Arg 764 recognizes a conserved guanine. Second, the two-finger expansion of ZF11 allows PRDM9c to recognize three-base-pair-longer sequences. A tryptophan in the additional ZF interacts with a conserved thymine methyl group. Third, an Arg-Asp dipeptide immediately preceding the ZF helix, conserved in two PRDM9a fingers and three PRDM9c fingers, permits adaptability to variations from a C:G base pair (G-Arg interaction) to a G:C base pair (C-Asp interaction). This Arg-Asp conformational switch allows identical ZF modules to recognize different sequences. Our findings illuminate the molecular mechanisms for flexible and conserved binding of human PRDM9 alleles to their cognate DNA sequences.
Organizational Affiliation:
From the Department of Biochemistry, Emory University School of Medicine, Atlanta, Georgia 30322.