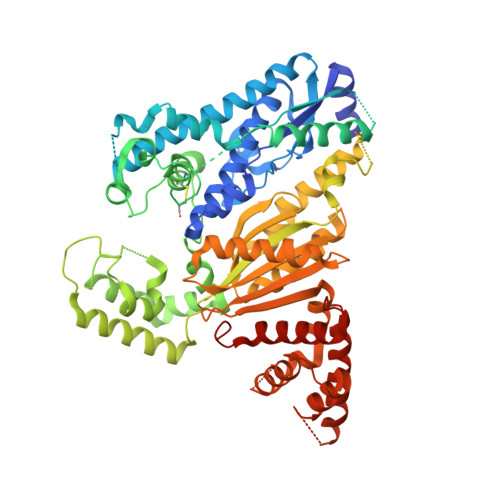
Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR-Cas systems
Zhu, X., Ye, K.(2012) FEBS Lett 586: 939-945
- PubMed: 22449983
- DOI: https://doi.org/10.1016/j.febslet.2012.02.036
- Primary Citation of Related Structures:
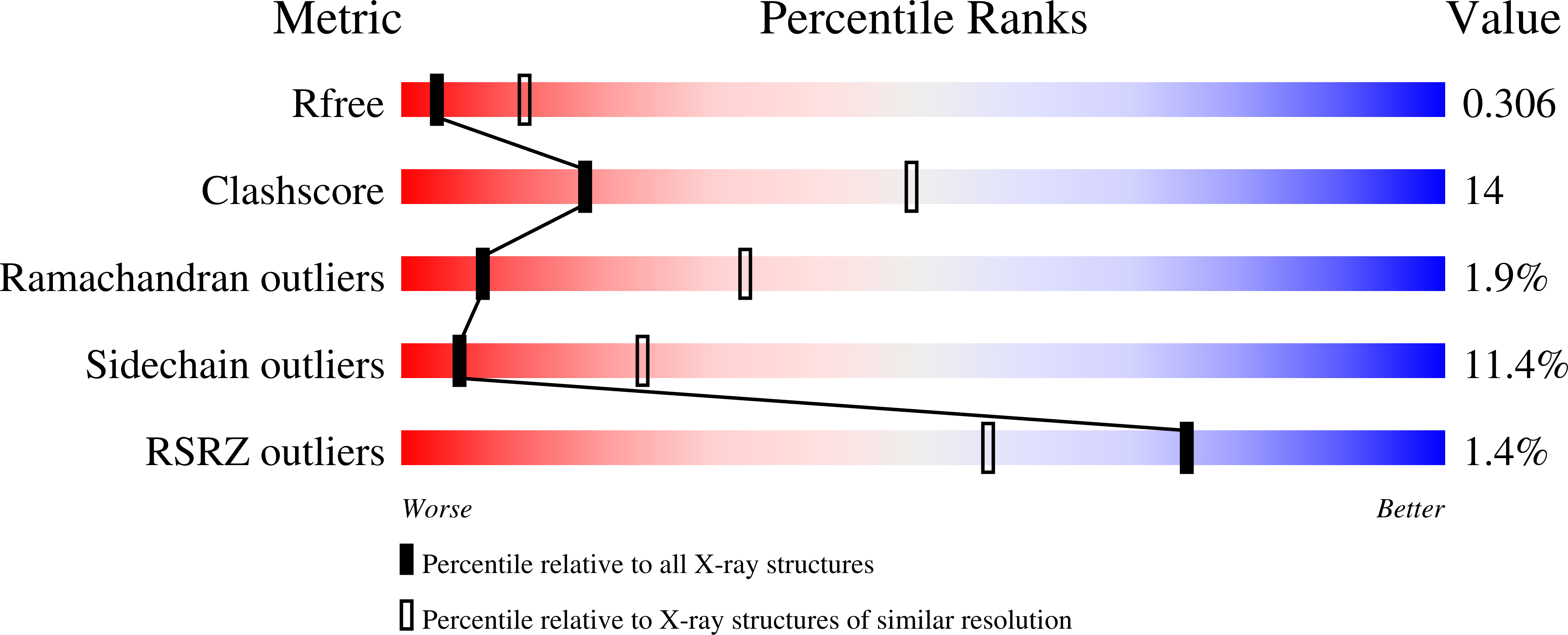
4DOZ - PubMed Abstract:
CRISPR RNAs (crRNAs) mediate sequence-specific silencing of invading viruses and plasmids in prokaryotes. The crRNA-Cmr protein complex cleaves complementary RNA. We report the crystal structure of Pyrococcus furiosus Cmr2 (Cas10), a component of this Cmr complex and the signature protein in type III CRISPR systems. The structure reveals a nucleotide cyclase domain with a set of conserved catalytic residues that associates with an unexpected deviant cyclase domain like dimeric cyclases. Additionally, two helical domains resemble the thumb domain of A-family DNA polymerase and Cmr5, respectively. Our results suggest that Cmr2 possesses novel enzymatic activity that remains to be elucidated.
Organizational Affiliation:
College of Life Sciences, Beijing Normal University, Beijing 100875, China.