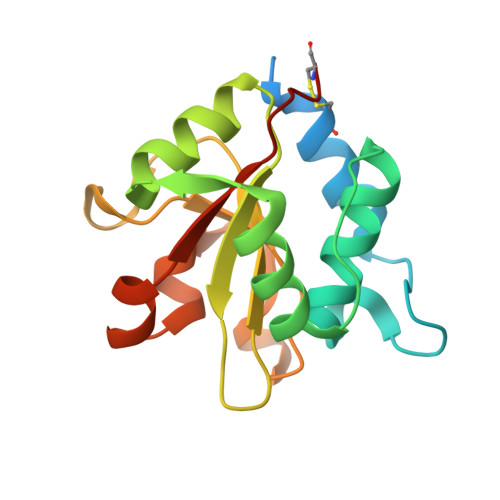
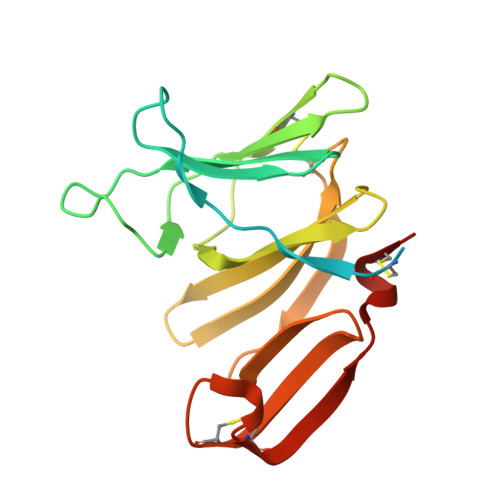
Structural insights into the Pseudomonas aeruginosa type VI virulence effector Tse1 bacteriolysis and self-protection mechanisms
Ding, J., Wang, W., Feng, H., Zhang, Y., Wang, D.C.(2012) J Biol Chem 287: 26911-26920
- PubMed: 22700987
- DOI: https://doi.org/10.1074/jbc.M112.368043
- Primary Citation of Related Structures:
3VPI, 3VPJ - PubMed Abstract:
Recently, it was identified that Pseudomonas aeruginosa competes with rival cells to gain a growth advantage using a novel mechanism that includes two interrelated processes as follows: employing type VI secretion system (T6SS) virulence effectors to lyse other bacteria, and at the same time producing specialized immunity proteins to inactivate their cognate effectors for self-protection against mutual toxicity. To explore the structural basis of these processes in the context of functional performance, the crystal structures of the T6SS virulence effector Tse1 and its complex with the corresponding immunity protein Tsi1 were determined, which, in association with mutagenesis and Biacore analyses, provided a molecular platform to resolve the relevant structural questions. The results indicated that Tse1 features a papain-like structure and conserved catalytic site with distinct substrate-binding sites to hydrolyze its murein peptide substrate. The immunity protein Tsi1 interacts with Tse1 via a unique interactive recognition mode to shield Tse1 from its physiological substrate. These findings reveal both the structural mechanisms for bacteriolysis and the self-protection against the T6SS effector Tse1. These mechanisms are significant not only by contributing to a novel understanding of niche competition among bacteria but also in providing a structural basis for antibacterial agent design and the development of new strategies to fight P. aeruginosa.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 10010, China.