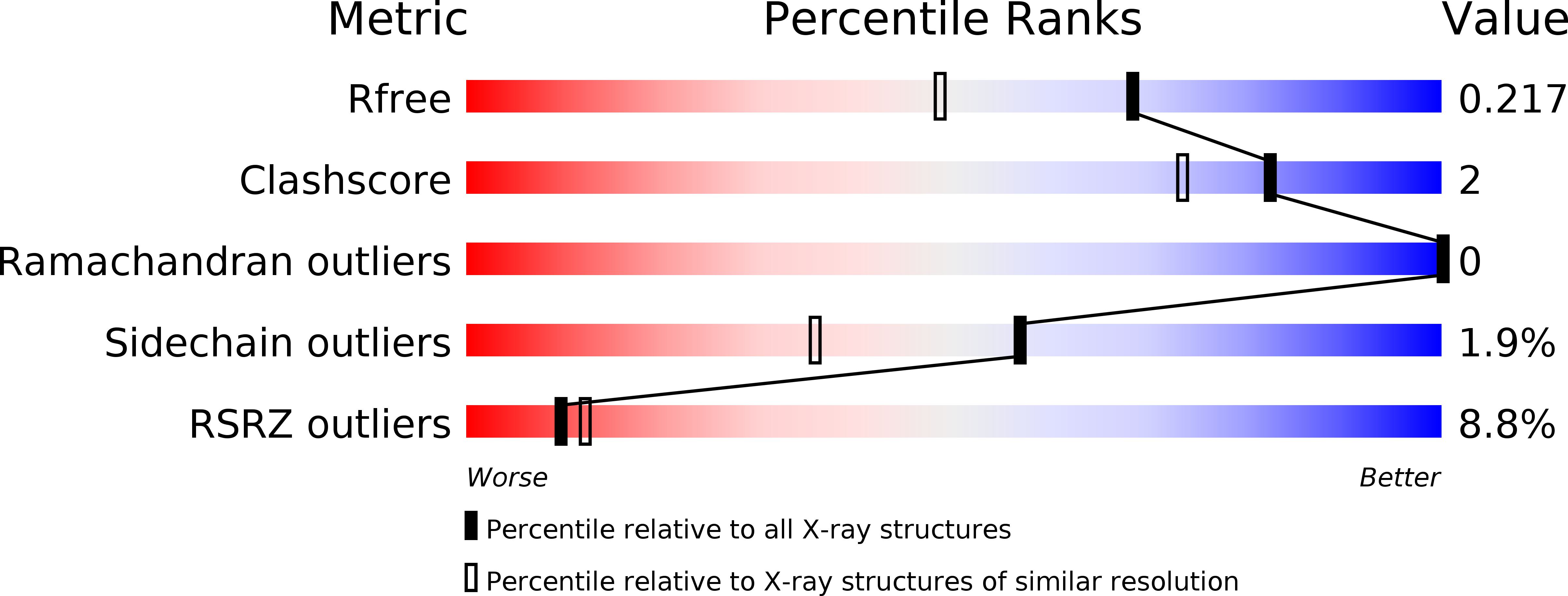
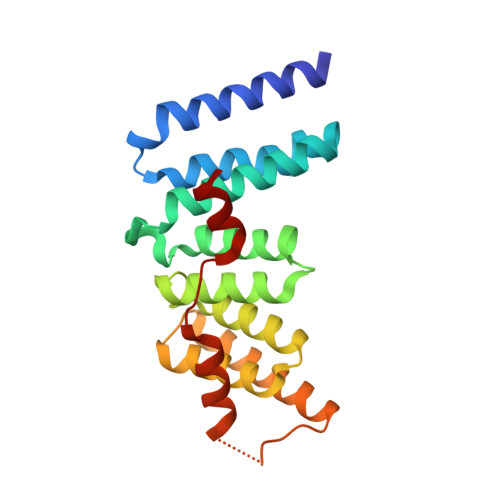
The Crystal Structure of the N-Terminal Region of BUB1 Provides Insight into the Mechanism of BUB1 Recruitment to Kinetochores.
Bolanos-Garcia, V.M., Kiyomitsu, T., D'Arcy, S., Chirgadze, D.Y., Grossmann, J.G., Matak-Vinkovic, D., Venkitaraman, A.R., Yanagida, M., Robinson, C.V., Blundell, T.L.(2009) Structure 17: 105-116
- PubMed: 19141287
- DOI: https://doi.org/10.1016/j.str.2008.10.015
- Primary Citation of Related Structures:
3ESL - PubMed Abstract:
The interaction of the central mitotic checkpoint component BUB1 with the mitotic kinetochore protein Blinkin is required for the kinetochore localization and function of BUB1 in the mitotic spindle assembly checkpoint, the regulatory mechanism of the cell cycle that ensures the even distribution of chromosomes during the transition from metaphase to anaphase. Here, we report the 1.74 angstroms resolution crystal structure of the N-terminal region of BUB1. The structure is organized as a tandem arrangement of three divergent units of the tetratricopeptide motif. Functional assays in vivo of native and site-specific mutants identify the residues of human BUB1 important for the interaction with Blinkin and define one region of potential therapeutic interest. The structure provides insight into the molecular basis of Blinkin-specific recognition by BUB1 and, on a broader perspective, of the mechanism that mediates kinetochore localization of BUB1 in checkpoint-activated cells.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, UK. victor@cryst.bioc.cam.ac.uk